Hi Viggo,
Thanks a lot for raising this issue! I think the problem is related to the base score of the gradient-boosted trees (GBRT) training. For more information https://stackoverflow.com/questions/47596486/xgboost-the-meaning-of-the-base-score-parameter
We need to read this constant value from the ONNX representation and add it to the output. I'll try to look at this in the coming days if I have some time. In the meantime, it seems that most GBRT algorithms use the mean of the training data as the base score. So for a quick fix, adding the mean to the output of the block should give the correct result.
import numpy as np
from sklearn.ensemble import GradientBoostingRegressor
from skl2onnx import convert_sklearn
from skl2onnx.common.data_types import FloatTensorType
import pyomo.environ as pyo
from omlt import OmltBlock
from omlt.gbt import GBTBigMFormulation, GradientBoostedTreeModel
import matplotlib.pyplot as plt
# Train simple model
x = np.array(range(101)).reshape(-1,1)
y = np.array(range(101))
model_gbt = GradientBoostingRegressor(n_estimators=5)
model_gbt.fit(x, y)
# Get predictions directly from model
y_pred = model_gbt.predict(np.array(range(100)).reshape(-1,1))
# Get predictions from pyomo formulation
y_opt = []
for i in range(100):
initial_type = [('float_input', FloatTensorType([None, 1]))]
model_onx = convert_sklearn(model_gbt, initial_types=initial_type)
m = pyo.ConcreteModel('Random GradientBoostingRegressor')
m.gbt = OmltBlock()
input_bounds = {0: (i, i)} # Forces the input to equal the input from the previous prediction
gbt_model = GradientBoostedTreeModel(model_onx, scaled_input_bounds=input_bounds)
formulation = GBTBigMFormulation(gbt_model)
m.gbt.build_formulation(formulation)
m.obj = pyo.Objective(expr=0)
solver = pyo.SolverFactory('cbc')
status = solver.solve(m, tee=False)
y_opt.append(m.gbt.outputs[0].value + np.mean(y))
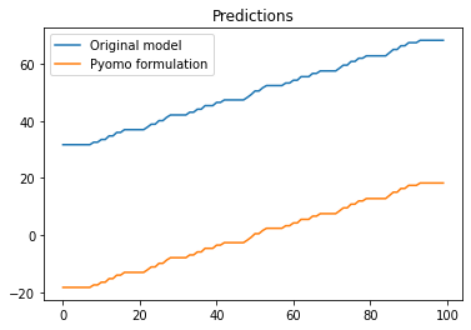
# Plot predictions
plt.plot(y_pred)
plt.plot(y_opt)
plt.title('Predictions')
plt.legend(['Original model', 'Pyomo formulation'])Output

Hi! I have an issue with the GradientBoostingRegressor not outputing the expcted values after being formulated as part of my pyomo model. The deviation seems to be a static offset, not random.
I'm pretty new to both pyomo and omlt, so this might be human error. I've also been looking for a list of supported models without luck, but I guess that could also be the issue.
I've attached an MRE below:
Output