Hi!
Thanks for the kind words and digging around the spectral interpolation jazz! :+1:
This is a currently known behaviour, and one we are not happy with! We are also working on it (more on this later).
In the current implementation of colour.spectral_to_XYZ definition we don't account for the differential distance dx when doing the operations. That's the ending 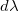 in those equations.
in those equations.
As a result the Luminance computed is different which should explain discrepancies you find in computing the CIE Lab values. That being said the colour should be almost the same (not accounting for the precision loss because of the different bin size and provided you normalise the values):
import colour
from colour.plotting import *
spd = colour.COLOURCHECKERS_SPDS['cc_ohta']['dark skin']
spd_c = spd.clone().interpolate(colour.SpectralShape(steps=10))
XYZ_a = colour.spectral_to_XYZ(spd) / 100
XYZ_b = colour.spectral_to_XYZ(spd_c) / 100
print('CIE XYZ:\n\t{0}\n\t{1}'.format(XYZ_a, XYZ_b))
print('CIE xy:\n\t{0}\n\t{1}'.format(colour.XYZ_to_xy(XYZ_a),
colour.XYZ_to_xy(XYZ_b)))
print('CIE Lab:\n\t{0}\n\t{1}'.format(colour.XYZ_to_Lab(XYZ_a),
colour.XYZ_to_Lab(XYZ_b)))
print('CIE Lab (Y Nornalised):\n\t{0}\n\t{1}'.format(colour.XYZ_to_Lab(XYZ_a / XYZ_a[1]),
colour.XYZ_to_Lab(XYZ_b / XYZ_b[1])))
spds_CIE_1931_chromaticity_diagram_plot((spd, spd_c))CIE XYZ:
[ 0.02386821 0.0199886 0.01118792]
[ 0.01193872 0.01000525 0.00557721]
CIE xy:
[ 0.43361472 0.3631338 ]
[ 0.43380111 0.36354738]
CIE Lab:
[ 15.48126316 10.02488061 6.58787558]
[ 8.99581624 7.93233569 4.98396404]
CIE Lab (Y Nornalised):
[ 100. 36.93899272 24.27455226]
[ 100. 36.81219815 24.51422506]
Now as I said, @MichaelMauderer and I are not happy about that and we are working on a complete overhaul of the spectral code. The BIG problem is that we actually can't just say: "Hey! Let's interpolate! \o/". Doing so implies that we know which interpolation type to use, etc... As we worked on that, we realised that we have opened an interesting can of worm and that it will also require a substantial amount of work.
So where we are now? Code wise, this is a prototype for the new spectral code: http://nbviewer.ipython.org/github/colour-science/colour-ramblings/blob/master/spectral_signal.ipynb We are only handling Spectrum object for now and need to discuss how we want to build the Spectra object. I got side-tracked those last months but I will get back to that in January.
The interpolation can of worm analysis draft is here: http://nbviewer.ipython.org/github/colour-science/colour-ramblings/blob/master/analysis_of_spectral_data_interpolation.ipynb
Voila! :)
First of all, a big thank you for creating this excellent python library for color research! Something like this was sorely missing and this is so well written!
While I was testing it, I think I may have stumbled across a bug in the spectral_to_XYZ() conversion. Currently, inside this conversion, if the shape of the SPD does not match the shape of color matching functions, the "missing" SPD wavelength data is being zero filled. In my opinion, this should be replaced by interpolation. I have done some testing using the color checker SPD data included in the library.
Values below are CIELAB calculated using spectral_to_XYZ then XYZ_to_Lab functions using D65 illuminant for "BabelColor Average" and "ColorChecker N Ohta" SPDs. (I have trimmed it to the first 5 patches only).
As you will notice, there is a big difference between the CIELAB values of Babel ColorChecker data versus the Ohta ColorChecker data, when zero filling the missing wavelengths, something I did not expect. I see that the Ohta CIELAB values are calculated from 5 nm data whereas the Babel values are calculated from 10 nm data. So I changed the zero filling to interpolate and they are much closer (I am aware that these are from different measurements so they cannot be the same).
Apologies for the long explanation. I would be happy to create a PR for this fix, let me know!
Thanks! Rohit Color Scientist