Hi @dugu9sword
Based on your error message, your star file is missing the 'rlnImageSize' column in the optics group.
The easiest thing to get around this would be to modify your star file. Optics groups are defined on the relion wiki and have something like five columns if my memory serves.
@fredericpoitevin I think I had run into this too, but haven't had time to work with cryoAI much : (
If I could suggest: This case could be easily caught using a dictionaries get method, which would return 'None' on key error. For some keys, like this one, the value is implicitly defined by the input data, so could be deduced by cryoAI.



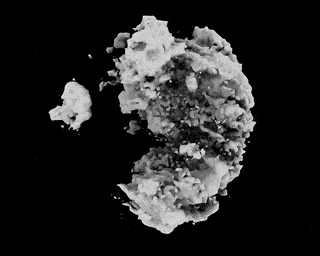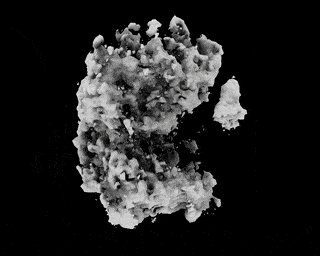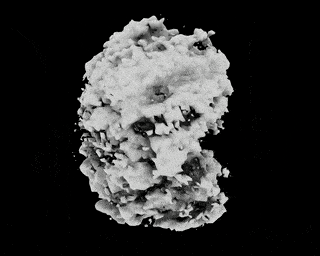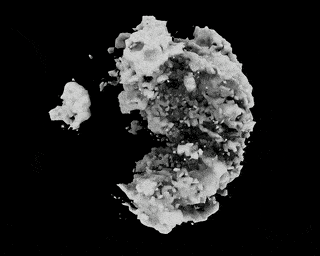
Hi,
I have downloaded the EMPIAR 10049 data following this link: https://github.com/zhonge/cryodrgn_empiar
But there are some questions:
.starfile, how can I resolve it?ctf.pkl?Thanks!