This seems to happen because GPyTorch defaults to fast computations. We recover the correct posterior if we disable preconditioned CG by wrapping the posterior in the following:
with gpytorch.settings.fast_computations(covar_root_decomposition=True, log_prob=True, solves=False):
# posterior calc...Interestingly enough the exact version is much faster in this case... (I am guessing CG is only really useful when N becomes truly big). Maybe these (default) "approximations" should be made more explicit if they can lead to weird behaviour?
PS: As far as I remember CG should actually give an exact solution provided enough steps. I will look into whether we can configurate it differently to reproduce the exact posterior...





🐛 Bug
I am trying to fit
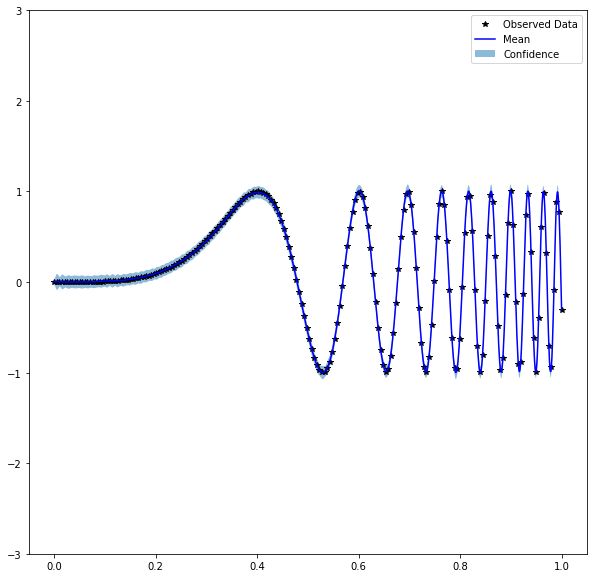
f(x) = sin(60 x⁴)with agpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel())kernel with many observations (N=1000). I have fixed the hyperparameters but the posterior variance seems to be much bigger than for GPy (see plots further down). We would expect it to approach zero everywhere for this many observations. Do you have a clue from where the instability could come?To reproduce
Code snippet to reproduce
Stack trace/error message
Expected Behavior
We expect similar behaviour to GPy:
But got:
System information
GPyTorch Version: 0.3.2 PyTorch Version: 1.0.1.post2 Computer OS: MacOS 10.14
Additional context
Code to reproduce the GPy plot: