@CaiziLee Thanks for pointing out the bugs. I guess the batchgenerators package has been updated, some of the functions have been moved to other places.
Looks like all the pre-trained models are not working in your case, have you checked the contrastive learning loss? My suggestion is to use a larger initial learning rate in contrastive learning like 0.1. Also, during fine-tuning, set the learning rate to the same as the train from scratch. Starting from 5e-5 seems to be too small.
Here is a contrastive learning loss example and a fine-tune result example of mine


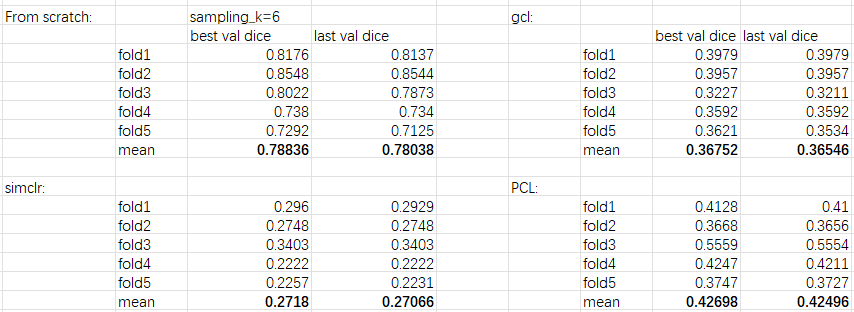
a very large gap between the reported results and mines, did I miss any important tricks? The details of my experiments are presented as follows, 1, fix two bugs: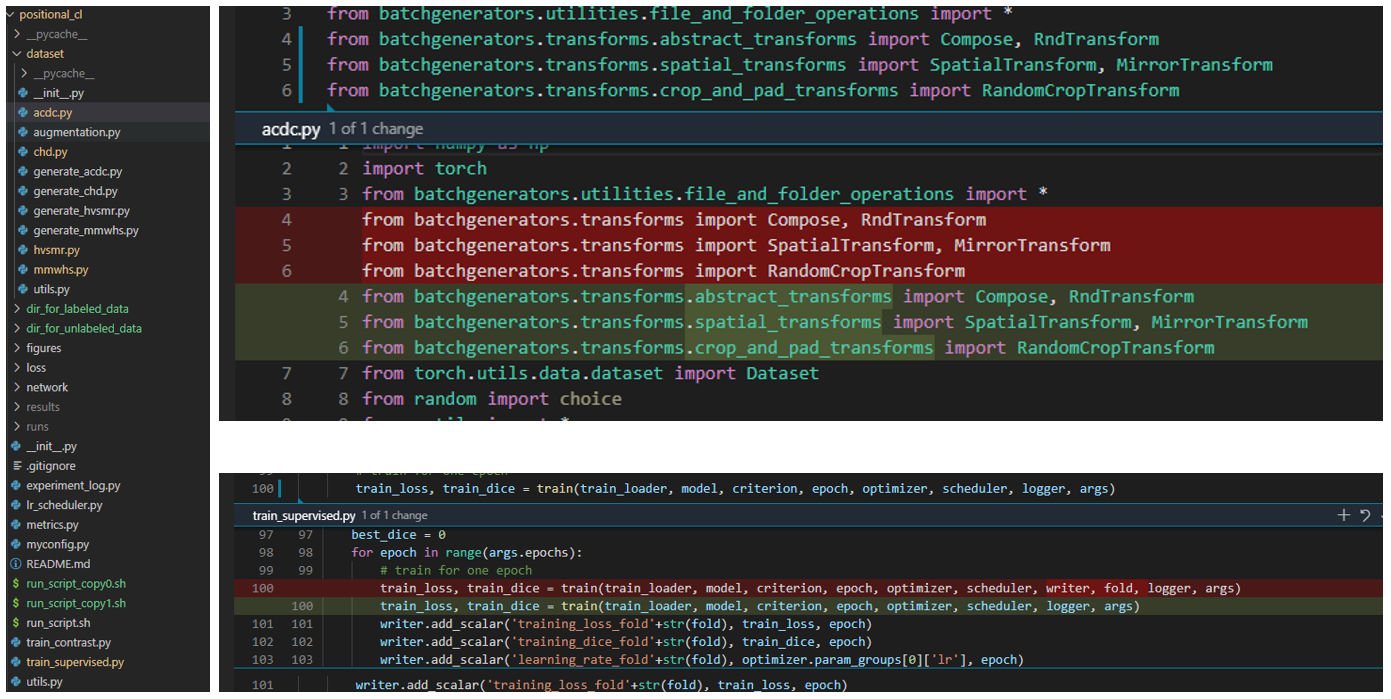
2, prepare acdc dataset via generate_acdc.py
3, prepare running scripts: (1) from scratch: python train_supervised.py --device cuda:0 --batch_size 10 --epochs 200 --data_dir ./dir_for_labeled_data --lr 5e-4 --min_lr 5e-6 --dataset acdc --patch_size 352 352 --experiment_name supervised_acdc_random_sample6 --initial_filter_size 48 --classes 4 --enable_few_data --sampling_k 6;
(2) contrastive learning: python train_contrast.py --device cuda:0 --batch_size 32 --epochs 300 --data_dir ./dir_for_unlabeled_data --lr 0.01 --do_contrast --dataset acdc --patch_size 352 352 --experiment_name contrast_acdc_pcl_temp01thresh035 --slice_threshold 0.35 --temp 0.1 --initial_filter_size 48 --classes 512 --contrastive_method pcl;
python train_contrast.py --device cuda:0 --batch_size 32 --epochs 300 --data_dir ./dir_for_unlabeled_data --lr 0.01 --do_contrast --dataset acdc --patch_size 352 352 --experiment_name contrast_acdc_gcl_temp01thresh035 --slice_threshold 0.35 --temp 0.1 --initial_filter_size 48 --classes 512 --contrastive_method gcl;
python train_contrast.py --device cuda:0 --batch_size 32 --epochs 300 --data_dir ./dir_for_unlabeled_data --lr 0.01 --do_contrast --dataset acdc --patch_size 352 352 --experiment_name contrast_acdc_simclr_temp01thresh035 --slice_threshold 0.35 --temp 0.1 --initial_filter_size 48 --classes 512 --contrastive_method simclr;
(3) finetuning: python train_supervised.py --device cuda:0 --batch_size 10 --epochs 100 --data_dir ./dir_for_labeled_data --lr 5e-5 --min_lr 5e-6 --dataset acdc --patch_size 352 352 --experiment_name supervised_acdc_simclr_sample6 --initial_filter_size 48 --classes 4 --enable_few_data --sampling_k 6 --restart --pretrained_model_path ./results/contrast_acdc_simclr_temp01_thresh035_2021-12-05_09-43-38/model/latest.pth;
python train_supervised.py --device cuda:1 --batch_size 10 --epochs 100 --data_dir ./dir_for_labeled_data --lr 5e-5 --min_lr 5e-6 --dataset acdc --patch_size 352 352 --experiment_name supervised_acdc_gcl_sample6 --initial_filter_size 48 --classes 4 --enable_few_data --sampling_k 6 --restart --pretrained_model_path ./results/contrast_acdc_gcl_temp01_thresh035_2021-12-04_03-46-35/model/latest.pth;
python train_supervised.py --device cuda:1 --batch_size 10 --epochs 100 --data_dir ./dir_for_labeled_data --lr 5e-5 --min_lr 5e-6 --dataset acdc --patch_size 352 352 --experiment_name supervised_acdc_pcl_sample6 --initial_filter_size 48 --classes 4 --enable_few_data --sampling_k 6 --restart --pretrained_model_path ./results/contrast_acdc_pcl_temp01_thresh035_2021-12-02_21-48-13/model/latest.pth;
4, experimental results(ubuntu16.04, pytorch1.9, NVIDIA 2080Ti * 2, Dice metric, take sample_k=6 as an example):