The samples in an MCMC chain are not independent (see https://emcee.readthedocs.io/en/stable/tutorials/autocorr/ for example) so it can be redundant to store many non-independent samples. thin=n takes the chain and returns every nth sample and you could set n to something like the integrated autocorrelation time without loss of statistical power. You will find that you get marginally different results because MCMC is a stochastic process and you'll see the Monte Carlo error.
General information:
Problem description: I'm wondering what does the parameter 'thin' exactly mean? I find for different thin value, the result is different. So I'm confused what value should I set to the parameter 'thin':
flat_samples = sampler.get_chain(discard=100, thin=?, flat=True)?let steps= 5000, walks=32, discard=100, and let
flat_samples = sampler.get_chain(discard=100, flat=True, thin=i)If I denote the size of flat_samples when thin= i as ' flat_samples| thin= i ', I found: flat_samples | thin=1 = (5000-100)32 =156800 flat_samples | thin=i = [ (5000-100)/thin] 32But I still do not understand the meaning of the parameter 'thin'. Why should one 'thin' a relatively big sample to a small sample?
Code to show my question:
By running the above code, I got a figure showing the relation between the value of thin and len(flat_samples):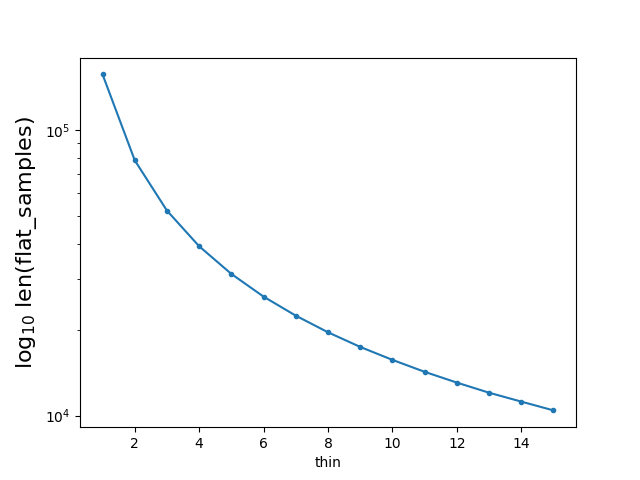
Then I made a corner plot by setting thin=1
I made another corner plot by setting thin=15, I found the figure is different: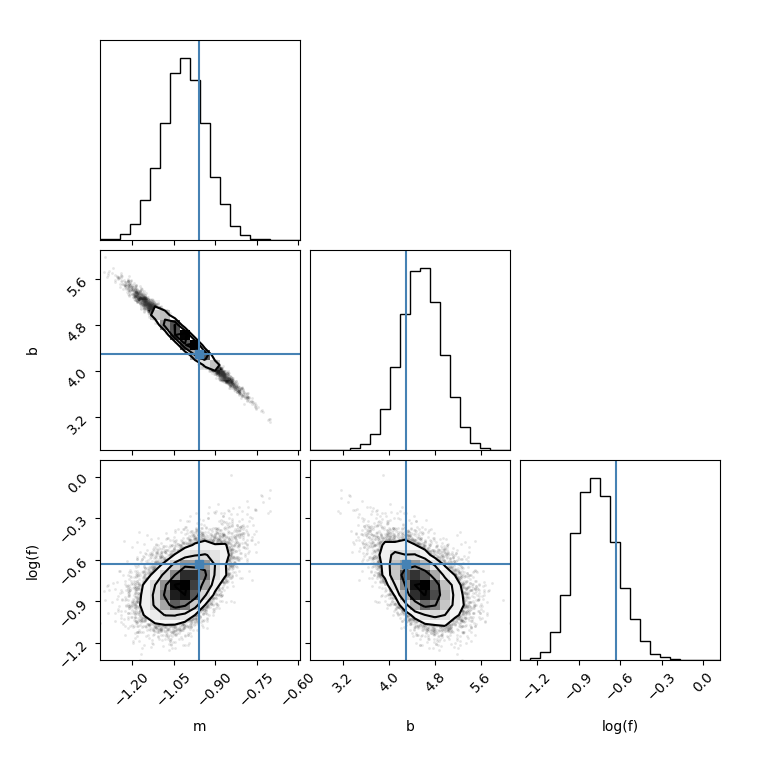
flat_samples = sampler.get_chain(discard=100, thin=15, flat=True)Then I run the following code, I found for different thin value, the results is also different.