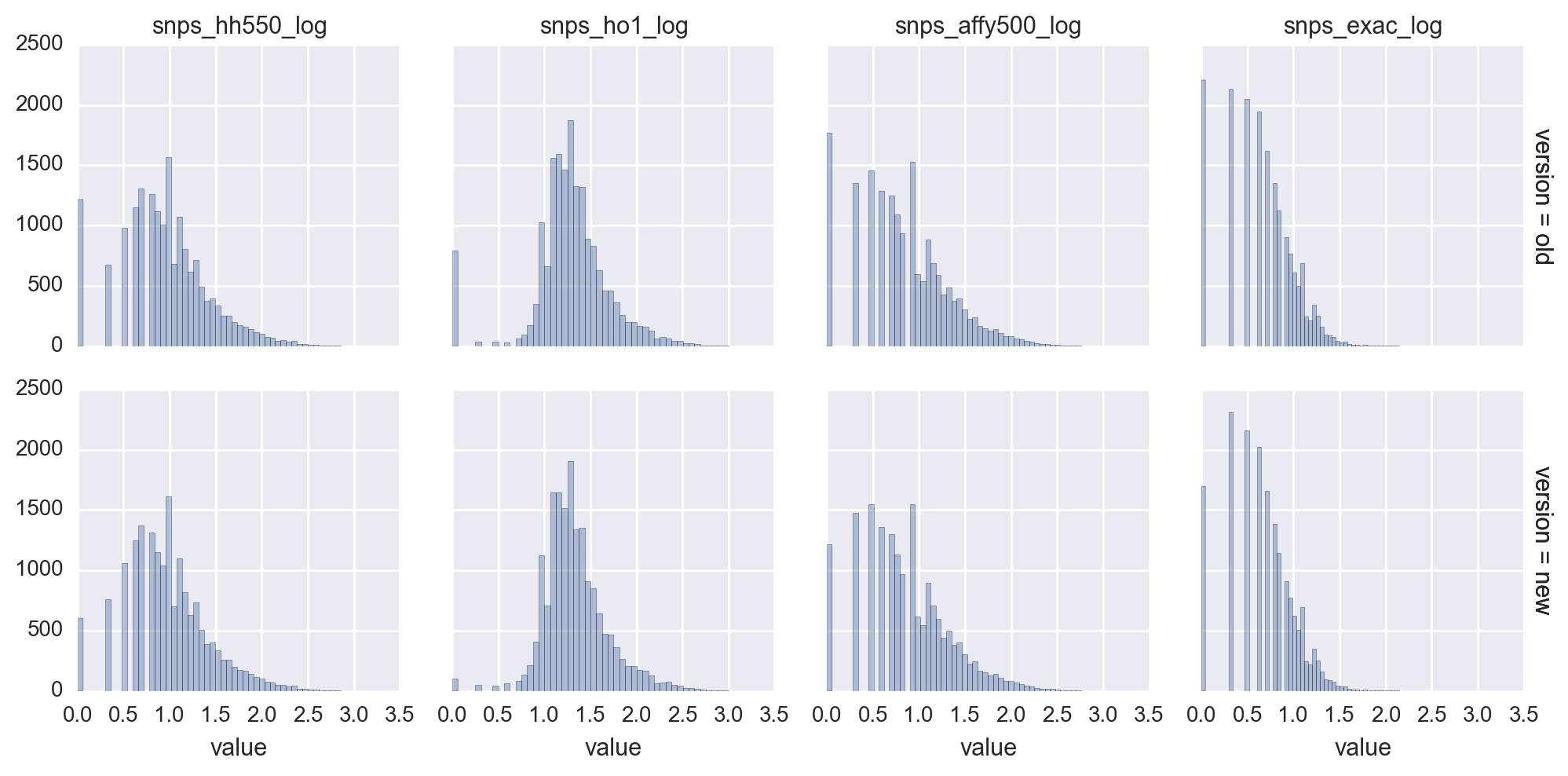
In 3007ac01d7cc4a753c277a4fd5a46771f58b5ce0, we confirm that HH550 SNP abundances for genes in chromosomes 1-22 did not change. We confirm their consistency.
For chrX, 89% of genes had at least one SNP on HH550 in the revised dataset. For chrY, only 20% of genes had at least one SNP.
I find it unlikely that changes to a small number of genes could have such a drastic effect.
HH550 through b795aa195a2c2f35bafb1e6b33c4819bff30d0a7
HH550 beginning on 5238adf66c9a2566e04ca81666fa6b4d47394fa1
Problem
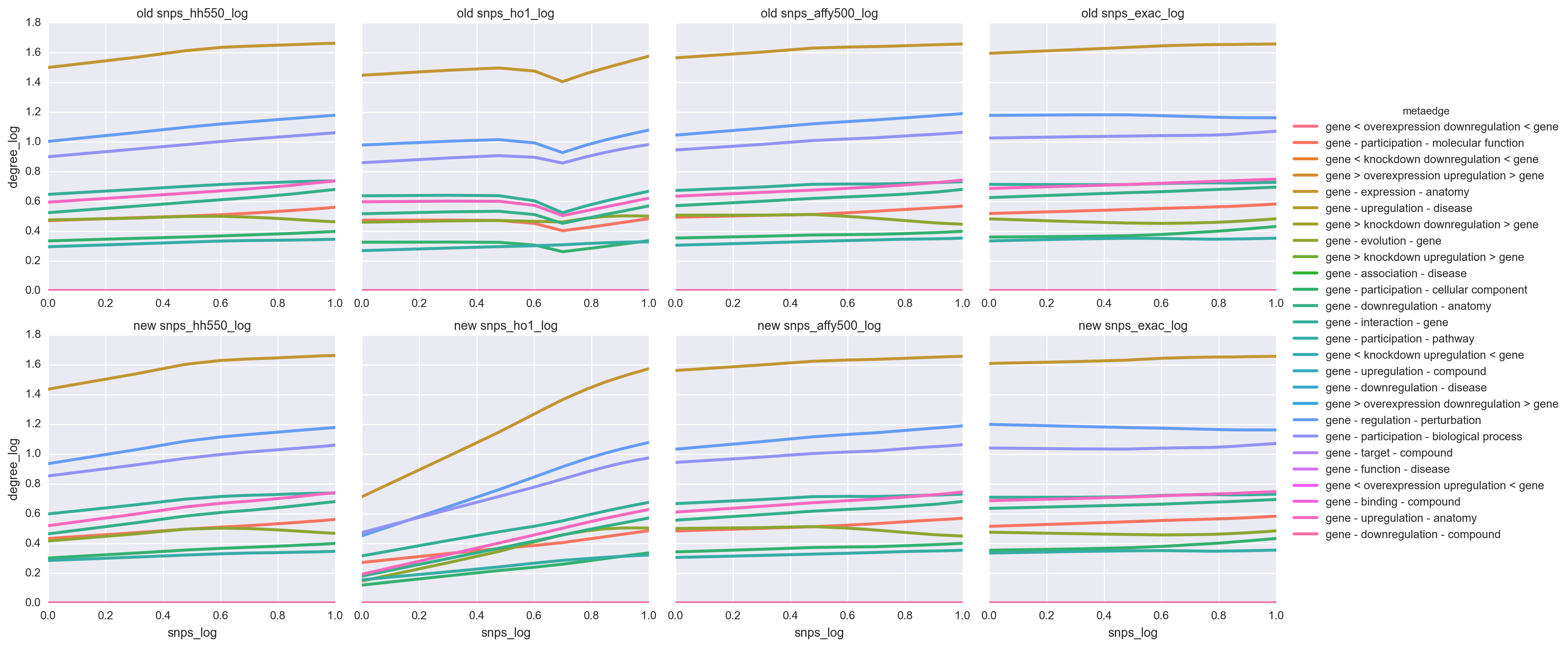
e783af22b660ad956cf9ea87994ece7e12ff3360 fixed a bug which was leading to all genes on chromosomes X and Y to receive 0 SNPs. The effect on 500K, HO1, and ExAC were minimal. However, the relationships for HH550 changed drastically with very low degrees being estimated for low SNP abundance.