model_parameters() does not report predictions (that should be in the modelbased package, but I guess not implemented yet). It reports the estimates, SE etc. from the summary. See:
library(metafor)
#> Loading required package: Matrix
#> Loading required package: metadat
#>
#> Loading the 'metafor' package (version 3.8-1). For an
#> introduction to the package please type: help(metafor)
library(parameters)
dat <- metadat::dat.bcg
datRR <- metafor::escalc(measure="RR",
data=dat,
ai=tpos, bi=tneg, ci=cpos, di=cneg)
res <- metafor::rma(data=datRR,
method = "REML",
mods = ~ alloc,
yi = yi, vi = vi,
slab = author)
summary(res)
#>
#> Mixed-Effects Model (k = 13; tau^2 estimator: REML)
#>
#> logLik deviance AIC BIC AICc
#> -10.3301 20.6602 28.6602 29.8705 36.6602
#>
#> tau^2 (estimated amount of residual heterogeneity): 0.3615 (SE = 0.2111)
#> tau (square root of estimated tau^2 value): 0.6013
#> I^2 (residual heterogeneity / unaccounted variability): 88.77%
#> H^2 (unaccounted variability / sampling variability): 8.91
#> R^2 (amount of heterogeneity accounted for): 0.00%
#>
#> Test for Residual Heterogeneity:
#> QE(df = 10) = 132.3676, p-val < .0001
#>
#> Test of Moderators (coefficients 2:3):
#> QM(df = 2) = 1.7675, p-val = 0.4132
#>
#> Model Results:
#>
#> estimate se zval pval ci.lb ci.ub
#> intrcpt -0.5180 0.4412 -1.1740 0.2404 -1.3827 0.3468
#> allocrandom -0.4478 0.5158 -0.8682 0.3853 -1.4588 0.5632
#> allocsystematic 0.0890 0.5600 0.1590 0.8737 -1.0086 1.1867
#>
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
model_parameters(res)
#> # Random Effects
#>
#> Parameter | Coefficient | SE | 95% CI | z | p | Weight
#> ----------------------------------------------------------------------------------
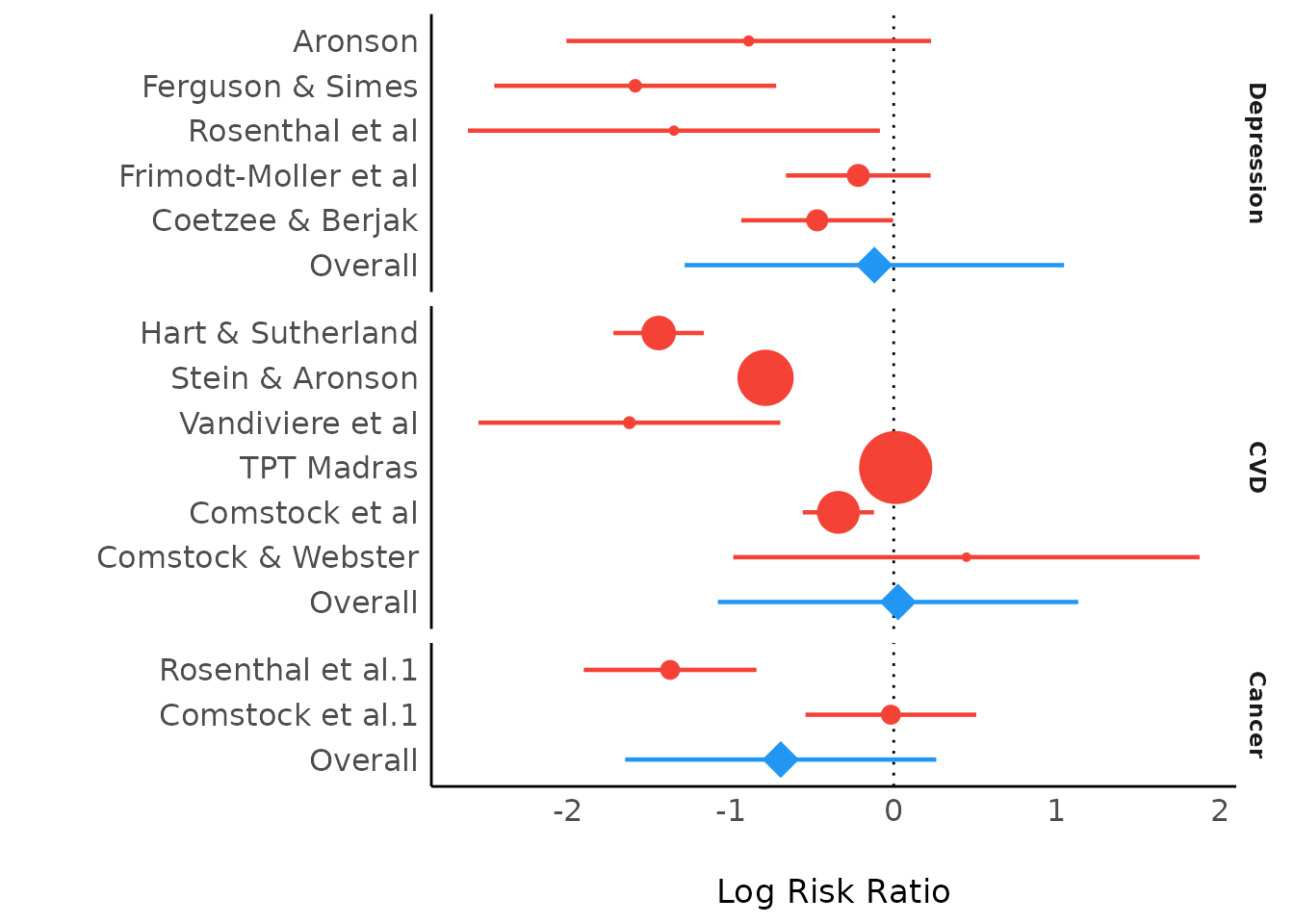
#> Aronson | -0.89 | 0.57 | [-2.01, 0.23] | -1.56 | 0.119 | 3.07
#> Ferguson & Simes | -1.59 | 0.44 | [-2.45, -0.72] | -3.59 | < .001 | 5.14
#> Rosenthal et al.1 | -1.35 | 0.64 | [-2.61, -0.08] | -2.09 | 0.036 | 2.41
#> Hart & Sutherland | -1.44 | 0.14 | [-1.72, -1.16] | -10.19 | < .001 | 49.97
#> Vandiviere et al | -1.62 | 0.47 | [-2.55, -0.70] | -3.43 | < .001 | 4.48
#> TPT Madras | 0.01 | 0.06 | [-0.11, 0.14] | 0.19 | 0.849 | 252.42
#> Coetzee & Berjak | -0.47 | 0.24 | [-0.94, 0.00] | -1.98 | 0.048 | 17.72
#> Overall | -0.45 | 0.52 | [-1.46, 0.56] | -0.87 | 0.385 |
#>
#> # alternate
#>
#> Parameter | Coefficient | SE | 95% CI | z | p | Weight
#> ------------------------------------------------------------------------------------
#> Frimodt-Moller et al | -0.22 | 0.23 | [-0.66, 0.23] | -0.96 | 0.336 | 19.53
#> Stein & Aronson | -0.79 | 0.08 | [-0.95, -0.62] | -9.46 | < .001 | 144.81
#> Overall | -0.52 | 0.44 | [-1.38, 0.35] | -1.17 | 0.240 |
#>
#> # systematic
#>
#> Parameter | Coefficient | SE | 95% CI | z | p | Weight
#> ----------------------------------------------------------------------------------
#> Rosenthal et al.2 | -1.37 | 0.27 | [-1.90, -0.84] | -5.07 | < .001 | 13.69
#> Comstock et al.1 | -0.34 | 0.11 | [-0.56, -0.12] | -3.05 | 0.002 | 80.57
#> Comstock & Webster | 0.45 | 0.73 | [-0.98, 1.88] | 0.61 | 0.541 | 1.88
#> Comstock et al.2 | -0.02 | 0.27 | [-0.54, 0.51] | -0.06 | 0.948 | 14.00
#> Overall | 0.09 | 0.56 | [-1.01, 1.19] | 0.16 | 0.874 |
#>
#> Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
#> using a Wald z-distribution approximation.Created on 2022-09-03 with reprex v2.0.2
The "Overall" rows correspond to the three estimates from the summary.



Hi,
It seems that
model_parameters()isn't calculating the correct point estimate (and 95% CI) for all subgroups in a metafor::rma() object. Below, I show thatpredict()andemmeans()yield the same results for all subgroups, whilemodel_parameters()only recovers the "alternate" subgroup correctly.Created on 2022-09-03 with reprex v2.0.2