- Could you share the SPM12 scripts you used in your article for generating the segmentation needed by brain2mesh?
We used the SPM GUI for segmenting the brains, nothing special. here are a couple of slides used in our training workshop back in 2017
Again, brain2mesh is not a segmentation tool, but a mesh generator processing pre-generated segmentations, so whatever best pipeline/parameters people use SPM or other segmentation tools, brain2mesh is expected to work with the output without any issue.
- Brain2mesh.m: if v2m is missing you ask for downloading brain2mesh, but you should ask for downloading iso2mesh
agreed and fixed https://github.com/fangq/brain2mesh/commit/de9d22edb782a60a9cabaaa8137de53679db5527
imfill/imdilate: Lots of people do not have access to the Image Processing toolbox (outside of North American universities), it would be good to provide alternatives. The Matlab functions imfill and imdilate are probably much more generic than the basic b/w morphological operations you need there. We try to have everything in Brainstorm working without any extra toolbox, for these ones I recoded only what I needed: mri_dilate.m, mri_fillholes.m
agreed again. replacing these dependencies is helpful for portability. In iso2mesh, I had implemented something like thinbinvol.m and thickenbinvol.m for similar purposes, but those only work for binary images (for replacing bwmorph). I do need something fast because I call those multiple times.
For my purposes, I bet convn() is enough for replacing imdilate, but I just did a quick comparison between convn(A, ones(3,3,3)) and imdilate(A, ones(3,3,3)), but I could not understand the difference. let me take a further look.
For imfill, I will do some test for the function you cited, with a quick look, I am not sure if it is capable of detecting holes.
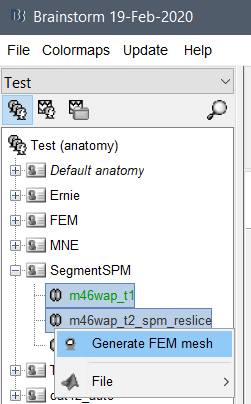


@fangq Following your suggestion, I'm checking how we could interface brain2mesh and brainstorm.
First few questions: 1) Could you share the SPM12 scripts you used in your article for generating the segmentation needed by brain2mesh? 2) Brain2mesh.m: if v2m is missing you ask for downloading brain2mesh, but you should ask for downloading iso2mesh 3) imfill/imdilate: Lots of people do not have access to the Image Processing toolbox (outside of North American universities), it would be good to provide alternatives. The Matlab functions imfill and imdilate are probably much more generic than the basic b/w morphological operations you need there. We try to have everything in Brainstorm working without any extra toolbox, for these ones I recoded only what I needed: mri_dilate.m, mri_fillholes.m
@tmedani @juangpc If you have solutions for this, feel free to share.