The original image will have an intensity associated value (assuming origin at zero) on 0.0, 1.0, and 2.0. If we resample to the desired spacing the image should have values on 0.0, 0.5, 1.0, 1.5, and 2.0. This results size 5.
I suppose this depends on whether you want the corners to be aligned or not. It's an implementation detail. I suppose that I implemented Resample to not have the corners (samples) aligned, but the "borders" of the pixels aligned.
The current implementation goes against the more digital-signal-processing choice, I suppose, as explained in A Pixel Is Not A Little Square, A Pixel Is Not A Little Square, A Pixel Is Not A Little Square! (And a Voxel is Not a Little Cube).
The default in PyTorch uses align_corners=False, which is what TorchIO does. If you think of pixels as little squares, or the Voronoi diagram of the pixel centers, the input and output should have similar bounds.
In practice, quantitative results seem to be very similar for both options.
Here's some code I just used to better understand the question:
import torch
import torchio as tio
tensor = torch.arange(3).reshape(1, 3, 1, 1).float()
image = tio.ScalarImage(tensor=tensor)
resampled = tio.Resample(0.5)(image)
resampled.data.flatten().tolist()
image_sitk = image.as_sitk()
resampled_sitk = resampled.as_sitk()
print('Original:')
for i in range(image_sitk.GetSize()[0]):
l, _ = image_sitk.TransformIndexToPhysicalPoint((i, 0))
print(-l, end=' ')
print()
print('Resampled:')
for i in range(resampled_sitk.GetSize()[0]):
l, _ = resampled_sitk.TransformIndexToPhysicalPoint((i, 0))
print(-l, end=' ')Output:
Original:
0.0 1.0 2.0
Resampled:
-0.25 0.25 0.75 1.25 1.75 2.25The "borders" of the images, considering that the sample is in the middle of the pixel, are (0 - 1/2, 2 + 1/2) = (-0.5, 2.5) for the original image and (-0.25 - 0.5/2, 2.25 + 0.5/2) = (-0.5, 2.5).
Here's a nice explanatory diagram to explain this idea (from this thread on the PyTorch forum):
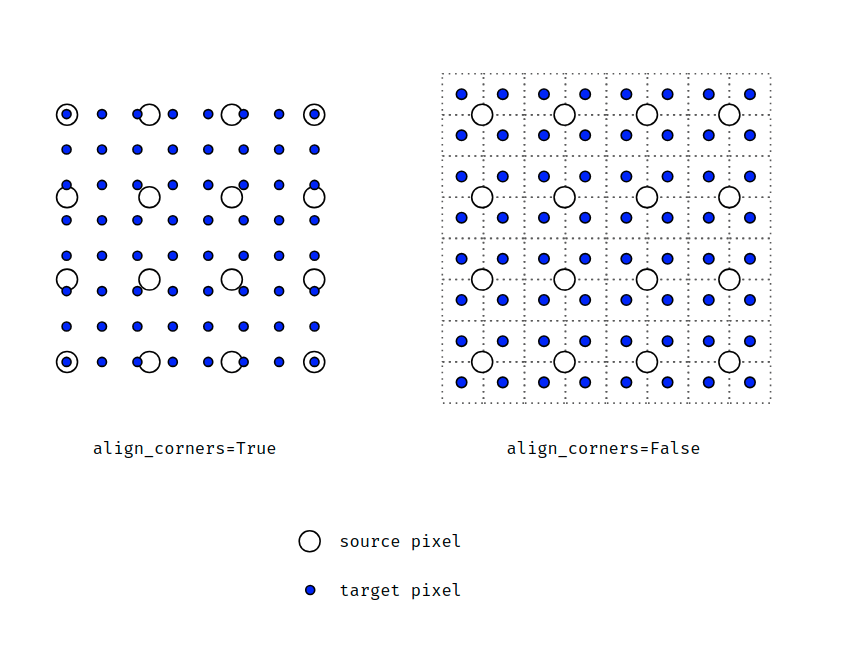
I hope some of that made sense.
Is this difference important for your use case? If it is, we could add an align_corners kwarg and try to mimic PyTorch's behavior.
Is there an existing issue for this?
Problem summary
After applying the resample transform to an image that has a spacing that is perfectly divisible by the new spacing there seems to be an extra voxel added. Let's take a single dimension with 1 mm spacing and size 3 as an example and a desired 0.5 mm spacing.
The original image will have an intensity associated value (assuming origin at zero) on
0.0,1.0, and2.0. If we resample to the desired spacing the image should have values on0.0,0.5,1.0,1.5, and2.0. This results size 5.Hence the following code should not raise an
AssertionError:The problem seems to be on the
get_reference_imagemethod which computes the new size as:I am not sure if the correct way to compute the new size is the following way:
which takes into account that the origin is shared among the images.
My issue stems from the fact that it should be possible (if the spacing is perfectly divisible) to upsample an image and recover it on the downsampling:
I also tried passing the affine and the shape:
But it didn't work either. I already tried it directly with
SimpleITKand it does recover the original image:Code for reproduction
Actual outcome
Shape is equal to
(1, 6, 6, 6).Error messages
No response
Expected outcome
Shape should be equal to
(1, 5, 5, 5).System info
No response