Your code is missing the part where you create the mesh. I guess you missed a line like
mesh=mt.createMesh(geom, quality=...The reason for the inaccuracy of the forward calculation is that the mesh is very coarse at the electrodes and it does not include them as nodes (not strictly needed but recommended). So either use
for electrode in scheme.sensors():
geom.createNode(electrode)or use mt.createParaMeshPLC(scheme) directly. Please usea large enough model boundary (yours ends with the electrodes) to improve the solution.
Please have a look at the ERT modelling examples in the example section of pygimli.org
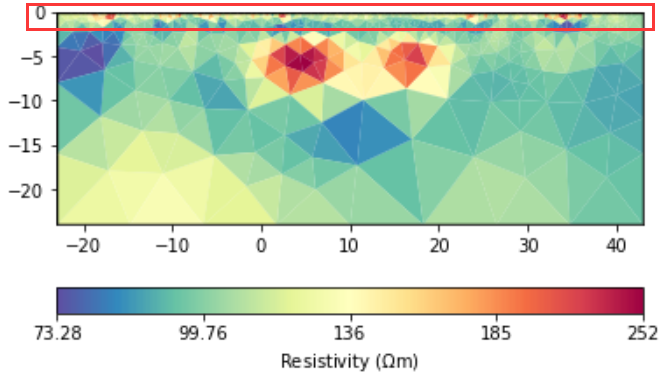
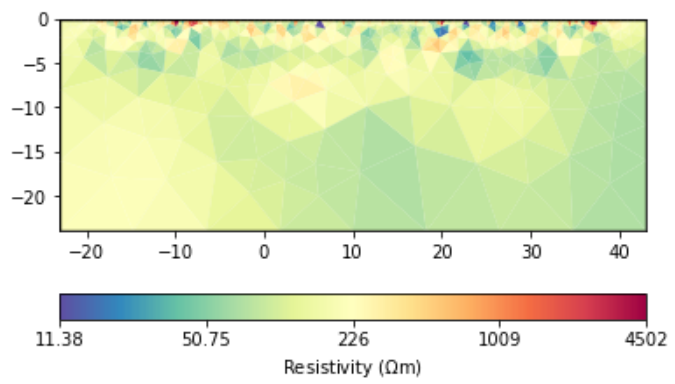
 If I expand the overall size like change the 'world' and refine the mesh ‘area=0.1’
If I expand the overall size like change the 'world' and refine the mesh ‘area=0.1’ 

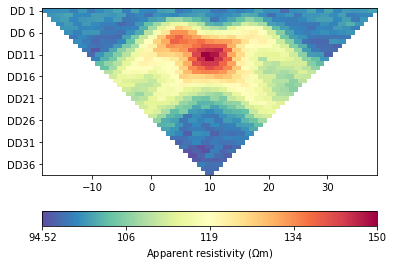
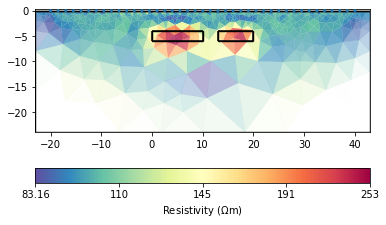

 The data generated by forward modeling are still the results of surface electrodes simulations.
The data generated by forward modeling are still the results of surface electrodes simulations.

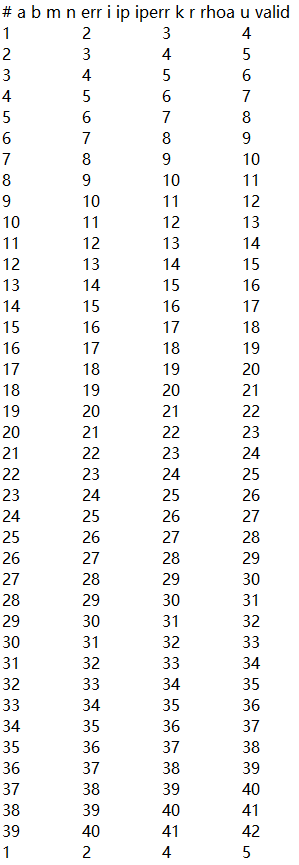
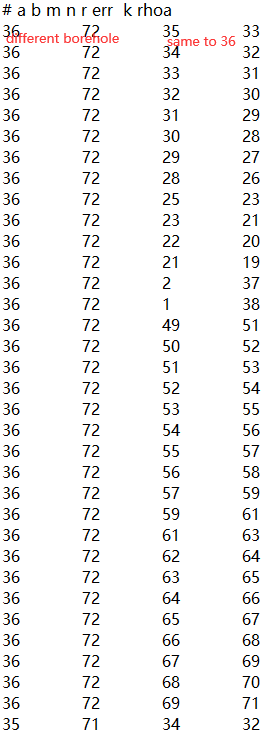
The forward and inversion results will have a large resistivity near the sensor. The larger the scale, the greater the resistivity. How to avoid abnormal values?
Other case:
 Looking forward to your reply! Thank you!
Looking forward to your reply! Thank you!