Adding @DaniallMasood here, he would know better for these questions and how the information should be structured but chipping in what I can:
- In the new data model, each biomarker entry represents a singular biomarker/disease connection whereas in the old model, you could have multiple diseases in the same biomarker entry. So for the top level
Generalsection, you could include the IDs, disease (condition) information, and best biomarker roles. Theassessed_biomarker_entityis within thebiomarker_componentarray because the new model is designed to more accurately capture panel/multi-component biomarkers. Soassessed_biomarker_entitywon't be in the general section. - The
biomarker_componentarray represents the different potential components of a biomarker whereas the instances (by our new definition) represent separate biomarkers. If there is only onebiomarker_componententry then the biomarker is a singular biomarker. If it has multiple components then it is a panel or multi-component biomarker meaning that it could be measured in a combination of assessed entity type's. Daniall would know what the (or if at all) theBiomarker Descriptionsection needs to be renamed. - I have no idea on this one, @DaniallMasood let us know.
- The reason why the
publicationsarray was renamed tocitationsis because we are accepting different types of citations that are not necessarily publications, for example patents and FDA certifications.
tst api response: https://api.tst.glygen.org/biomarker/detail/AA4686-11?query={"paginated_tables":[{"table_id":"publication","offset":1,"limit":200,"sort":"date","order":"desc"}]}
Production api response: https://api.glygen.org/biomarker/detail/A0001?query={"paginated_tables":[{"table_id":"publication","offset":1,"limit":200,"sort":"date","order":"desc"}]}
Left is tst api response and right is from production.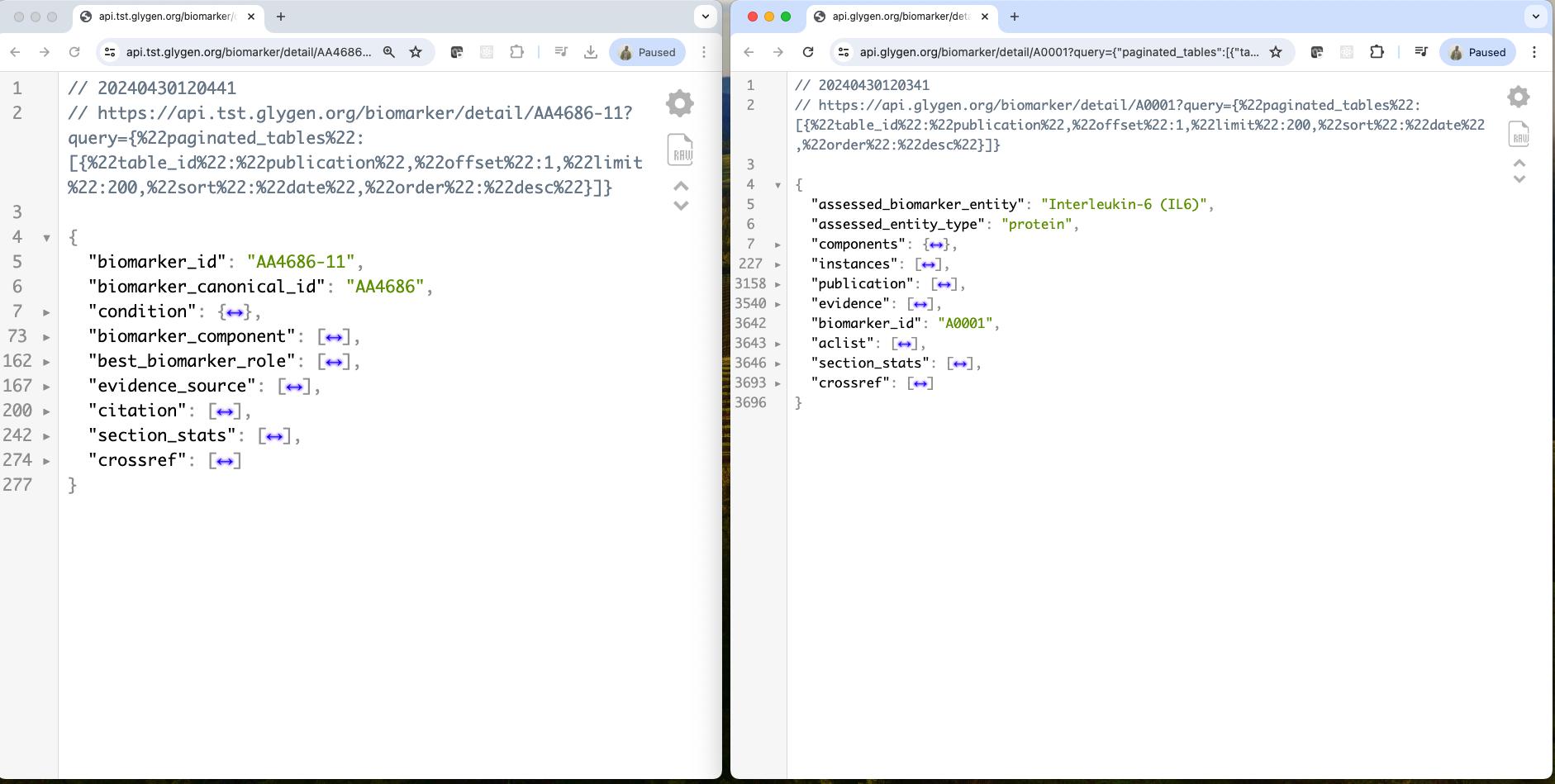
Old GlyGen interface https://www.glygen.org/biomarker/A0001
Few questions:
We have Five sections:
General New data model doesn't have something like assessed_biomarker_entity at top level of api response object.
Biomarker Description In the old data model data from instances":[] is mapped in Biomarker Description.
New data model doesn't have something like "instances":[] so do we need map data from biomarker_component:[] here? It looks like all fields are not there and structure is different.
Components In case of old data model components->protein and components->glycan is mapped to Glycan and Protein tables in Components section.
New data model doesn't have something like "components":{} so do we need map data from biomarker_component:[] here?
Cross References crossref:[] in both old and new data model.
Publications citation:[] array from new data model. publication:[] array from old data model.