I repeated the model after changing the rw1 formula from ~0 to ~1. I worried that fitting a zero intercept here may have forced the predictions to instead of using the current state as in
. The unexpected behavior (excess 0.5) persists despite this change.
brief description of the model
I'm attempting to implement a pure random walk model in which the underlying data generating process is described by) where y is a proportion between [0, 1]. The underlying state, y, is observed through counts of successes and failures using a logit link.
where y is a proportion between [0, 1]. The underlying state, y, is observed through counts of successes and failures using a logit link.
description of the unexpected behavior
The problem is that when I forecast from model fit with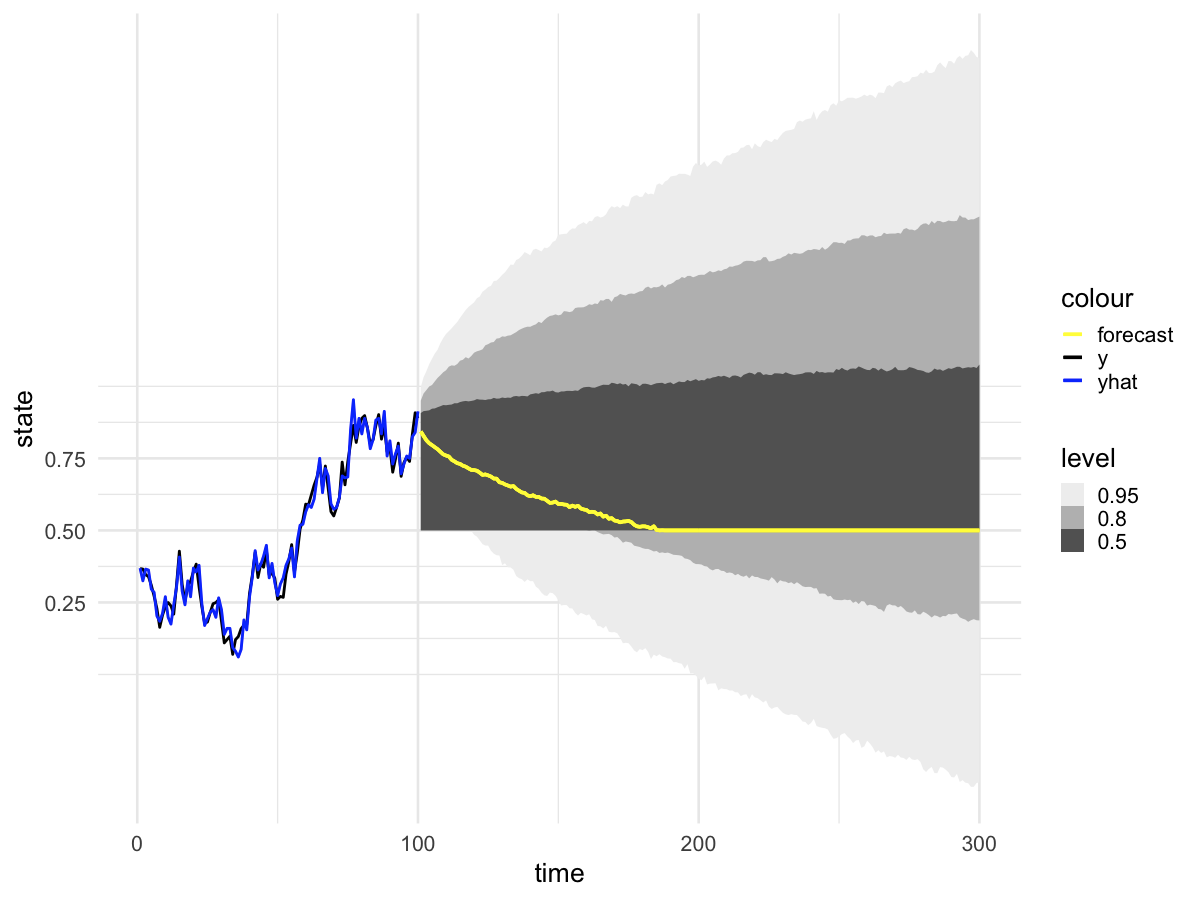
walker_glm(distribution = "binomial"), the prediction intervals behave strangely. Specifically, there is a very large excess of predictions at exactly 0.5 (corresponding to zero on the logit scale). The remainder of the posterior predictive distribution aligns with my expectations. We can visualize this problem in the posterior predictive intervals,and in the posterior predictions at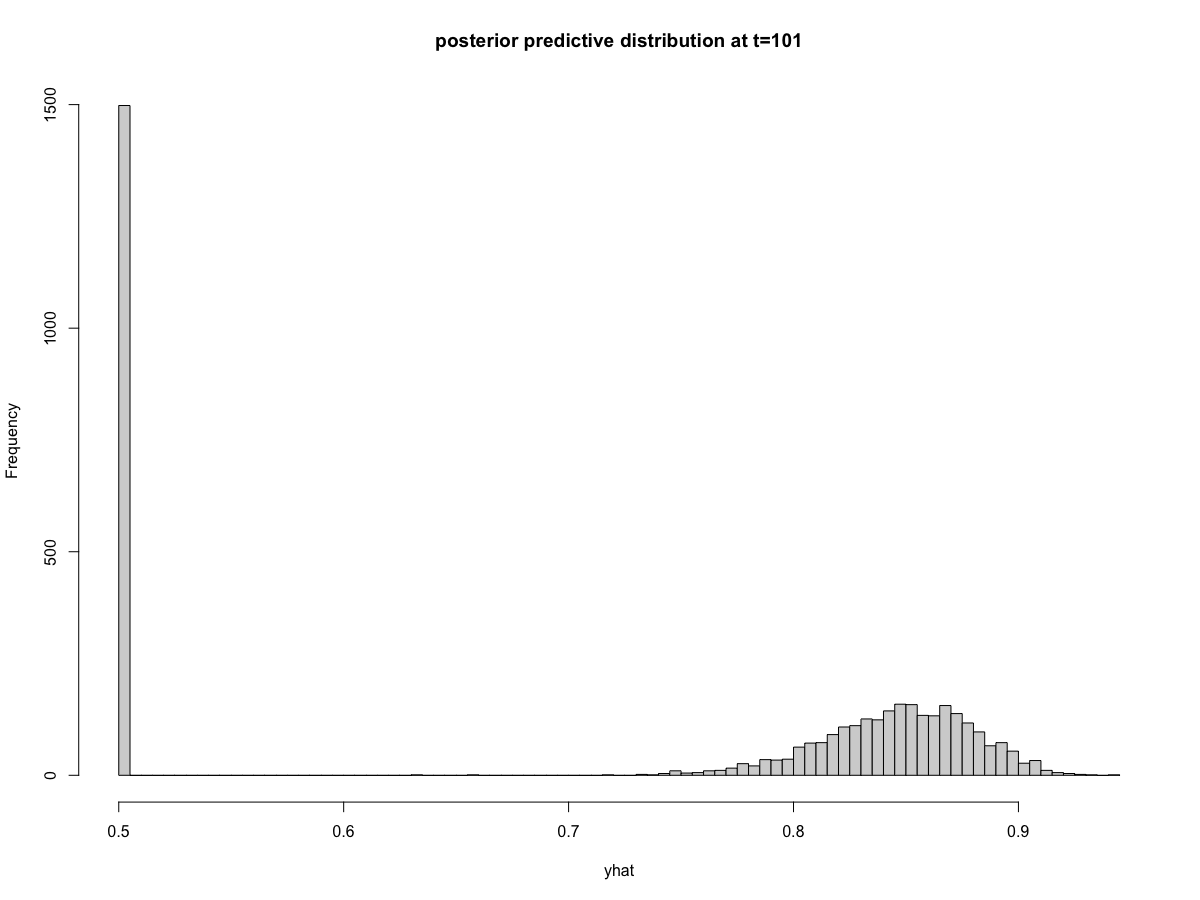
time=101and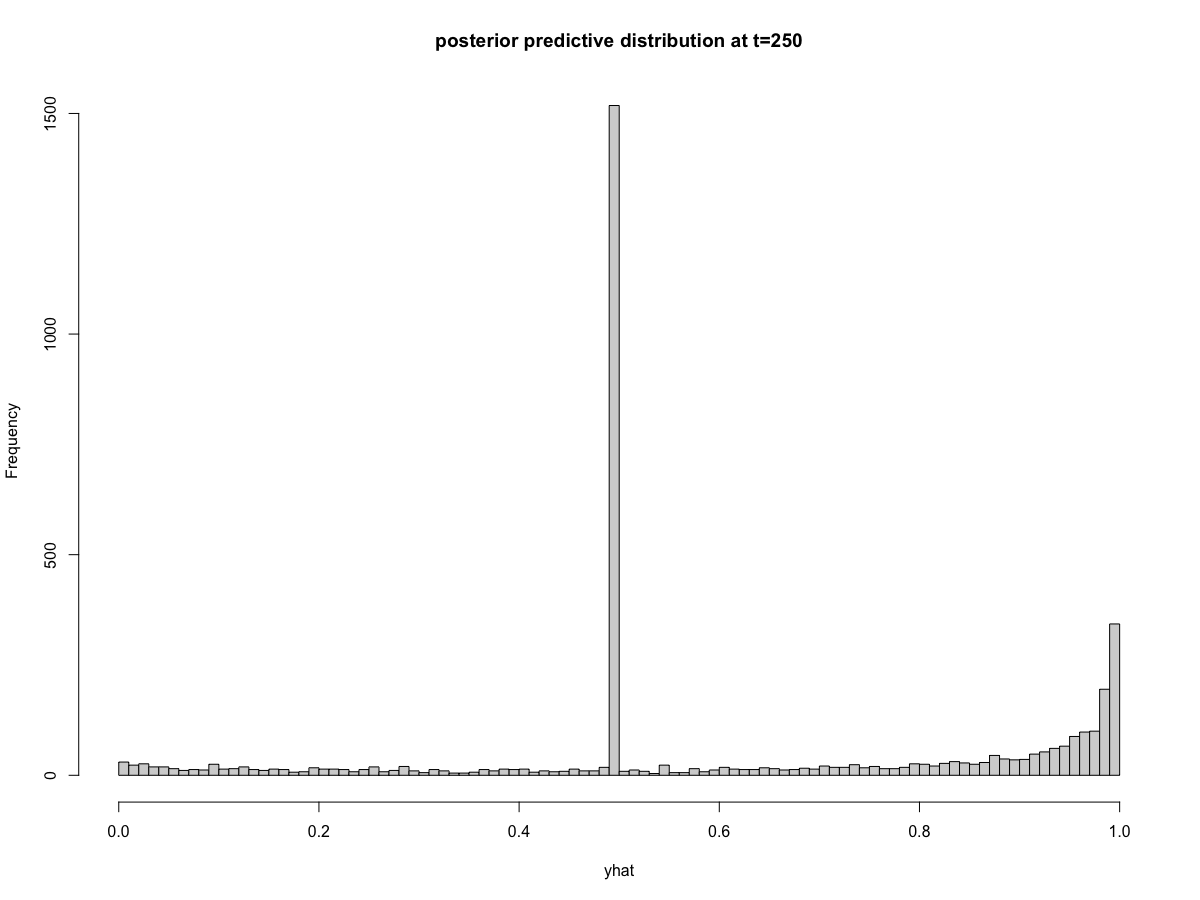
time=250potentially important clue
I noticed that all timepoints appear to have the same exact number of posterior predictions that equal 0.5 (again, this is the same as saying zero on the logit scale). You can see this in the histograms above where 1498 draws equal 0.5 at both
t=101andt=250(the exact count will vary depending on the random seed). The reprex below provides code that shows this is true across all time points. I can't think of a mechanism that would cause consistency of posterior draws across every time point.reprex
We can simulate
nt=100time steps of the true datay, the number of observed successessuccout ofnobstrials, and the estimated state on the probability scaleyhatas:We can then fit the model using
walker_glm()with a very weak prior on the value of yhat at time=0, and confirm that the parameters are correctly recovered:Finally, make the forecast for times 101 to 300:
Thank you, @helske, for your work on this package to make state space modeling intuitive and efficient.