This function was used to "flip" the alignments, basically pretending that the alignment was done on the reverse complement. In my example, it was used to go from
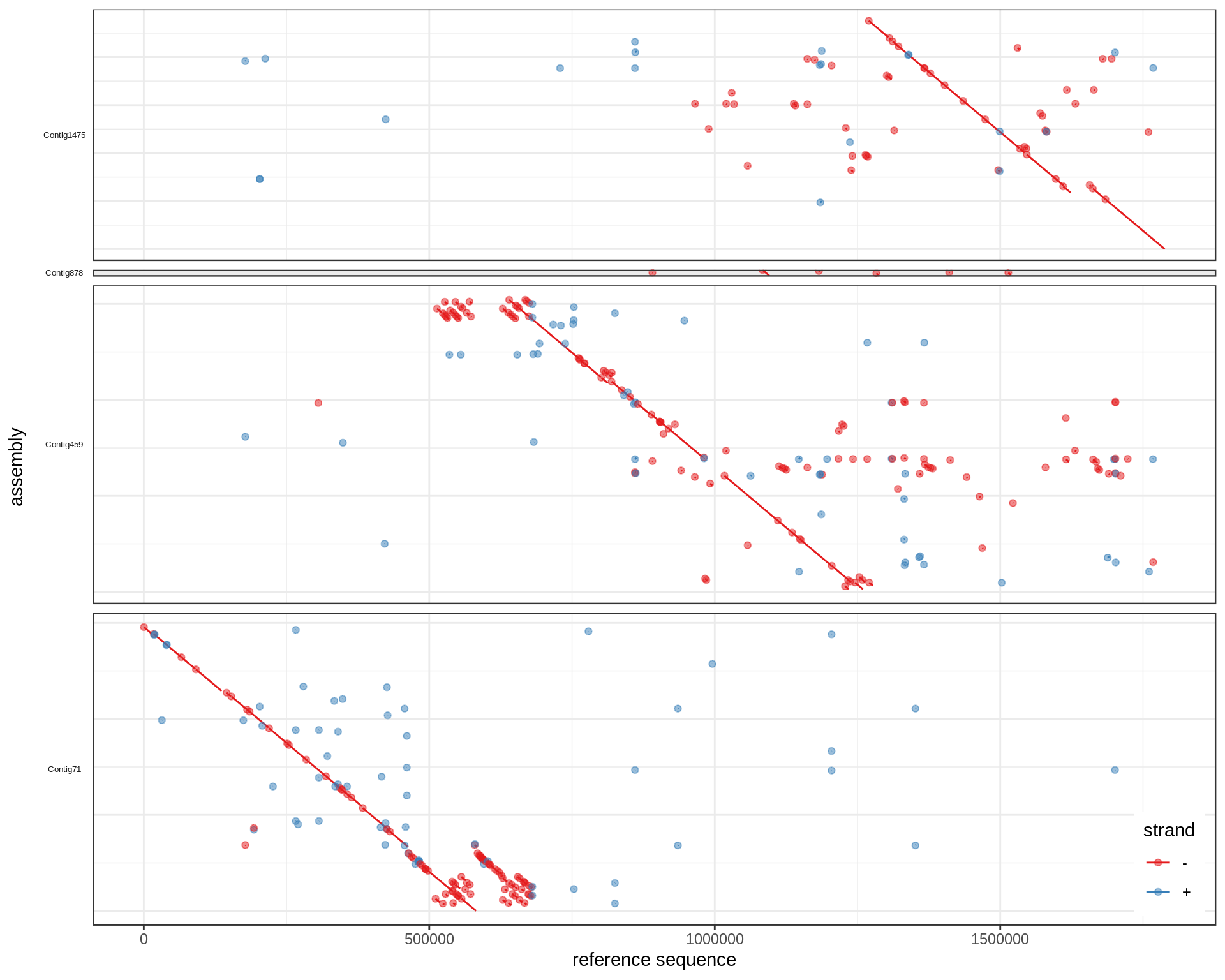
to
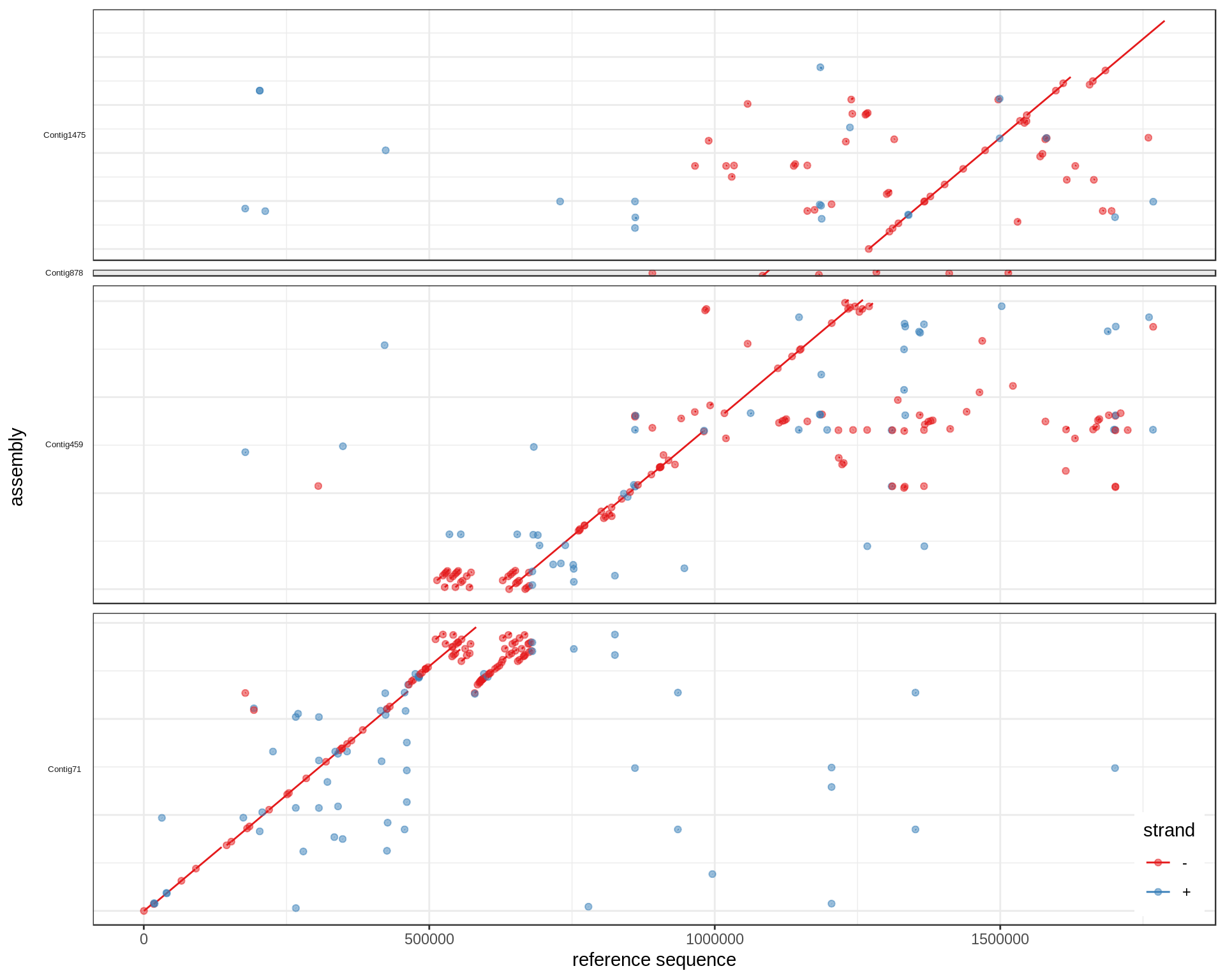
When a contig looks like it should be "flipped" (most bases aligned on - strand), the coordinates are transformed from qs to L-qs, where L is the length of the contig. For example, if an alignment block spans [80-90] of a 100bp contig, it would correspond to [10-20] of its reverse complemented sequence.
Of note, I didn't have the contig length easily accessible in that function, so I ended up using the maximum value of all the qe/qs instead. Not exactly ideal but fine for visualization purpose, esp. as those graphs don't have numbers on the y-axis.
I am using your code from here:
https://jmonlong.github.io/Hippocamplus/2017/09/19/mummerplots-with-ggplot2/
After using your function diagMum() the values of qs and qe are changing
For example, by writing only the first 6 columns, I have this before diagMum()
And, I have this after diagMum
Sorry if the answer is simple, but I don't understand what the function is doing and why the coordinates in the query are changing,
Many thanks.