Ok, I figured out a better solution to figure out the mean vector mean_vec. This uses the scatter matrix approach from Directional Statistics (Jupp and Mardia, 2000, pp.165).
import scipy.linalg as la
import numpy as np
# strikes, dips are variables containing orientations for the distribution of poles
lons, lats = mpl.stereonet_math.pole(np.mod(strikes,360),(dips))
x, y, z = mpl.stereonet_math.sph2cart(lons, lats)
xyz = np.hstack((x,y,z))
xyz = np.array(xyz).T
# xbar is the sample mean vector. This won't be accurate for vectors that point in opposite directions.
xbar = xyz.mean(axis=1)
# xobar is the sample mean direction
Rbar = np.linalg.norm(xbar)
xobar = xbar/Rbar
xobar = xobar.reshape(3,1)
# n is number of poles in the distribution
n = xyz.shape[1]
# initialize the scatter matrix T
T = np.zeros((3,3))
# Equation 9.2.10 from Directional Statistics (Jupp and Mardia, 2000, pp.165)
for i in range(n):
T += (xyz[:,i].reshape(3,1)).dot((xyz[:,i].reshape(3,1)).T)
T = T/n
# Mean vector of distribution is one of the eigenvectors of the scatter matrix T
eigs = la.eigh(T)
mean_vec = eigs[1][:,2]
mean_pole = mpl.stereonet_math.vector2pole(mean_vec[1],mean_vec[2],-1*mean_vec[0])
### print results to test
calculated_strike = mean_pole[0]
calculated_dip = mean_pole[1]
print(calculated_strike)
print(calculated_dip)
Hi Joe, The find_fisher_stats, fisher_stats, and mean_vector functions don't seem to be calculating Fisher k (kappa) or the mean vector of a group of poles correctly where the group of poles includes orientations which cross a dip=90°.
For example, the Fisher K and mean vector for this group of poles is correct. Calculated mean dip is 50°, Fisher K is 50.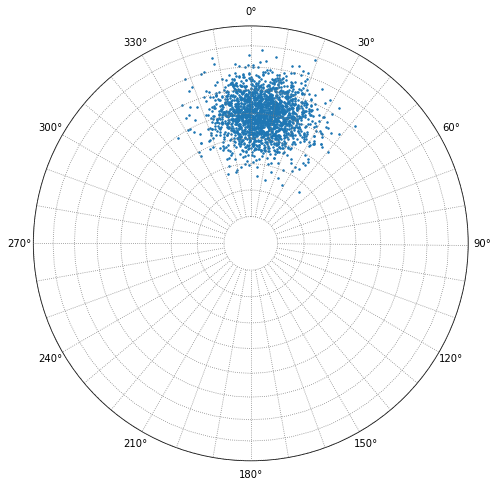
But, for this example the distribution has an actual Fisher k value of 25 and a mean dip of 85°, but the calculated kappa is 1.6 and the calculated mean dip is 62°.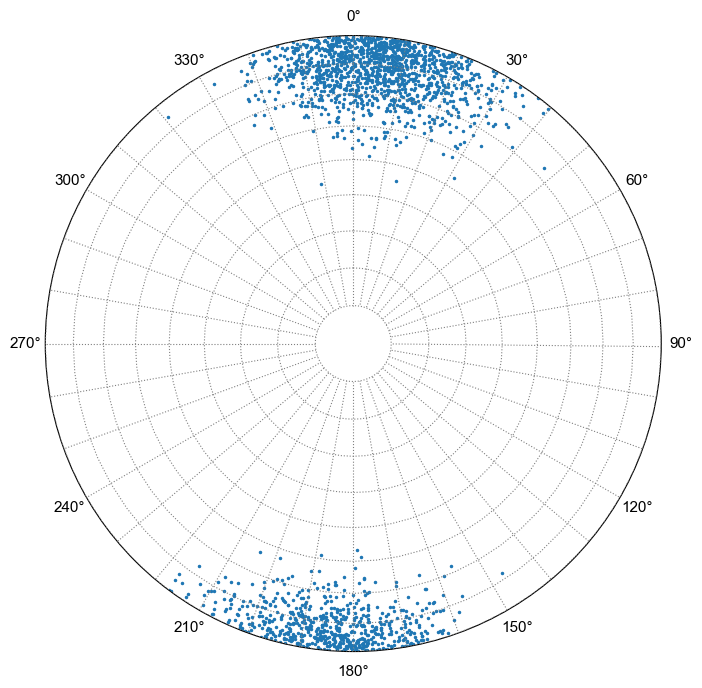
I think the issue is the lines of code where you take the mean of the vectors. xyz = np.vstack(xyz).T mean_vec = xyz.mean(axis=0)
When the largest eigenvalue flips direction, just taking the mean gives you a bad result.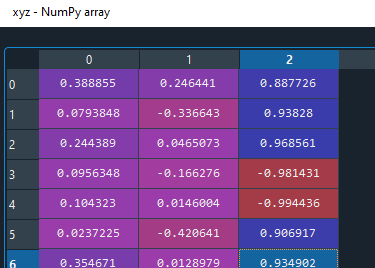
As a hack/workaround, I think you can do something like flip the normals so they all point in the same direction: le = np.argmax(np.max(np.abs(xyz), axis=0)) # largest eigenvalue is the one we should normalize the others by cond = xyz[:,le]<0 xyz = np.where(cond[:,None],xyz*-1,xyz)
But I bet you can come up with a better solution. This library is awesome, thanks for all your work on it!