It seems that adding an extra step after the classifier is not allowed?
Yes, this is a SCIKIT-LEARN LIMITATION (not a SkLearn2PMML limitation) - "a pipeline can contain at most one estimator (aka model) object, and if present, it must be located in the final position".
How can I implement adding extra rules after the classifier?
You can use PMMLPipeline.predict_transform(X), PMMLPipeline.predict_proba_transform(X), etc post-processing methods. IIRC, you've done it before.
Alternatively, if you wrap your estimator object into sklego.meta.EstimatorTransformer object, then it becomes a transformer (implements fit_transform(X) instead of fit_predict(X, y)), and the above limitation is effectively cancelled. That is, you can have multiple estimator-disguised-as-transformer objects in your pipeline, plus they can appear in positions other that the final position.
Is it impossible to achieve?
It is impossible using canonical Scikit-Learn pipelines.
But if you're open to using 3rd-party extension packages, and doing some thinking for yourself, it's easy-peasy.
Here's the thing, I'm building a machine learning pipeline using PMMLPipeline
Because I need to correct the prediction probability according to the value of a certain column after the classifier predicts.
For example, I have 12 columns of data, I need to use the first 11 columns for prediction, and then correct the prediction result according to the value of the 12th column (if the twelfth column is greater than 0, correct the prediction result to 0)
So my code is written as follows
But it prompts the following error
It seems that adding an extra step after the classifier is not allowed?
How can I implement adding extra rules after the classifier? Because I noticed that the result after the classifier seems to be an array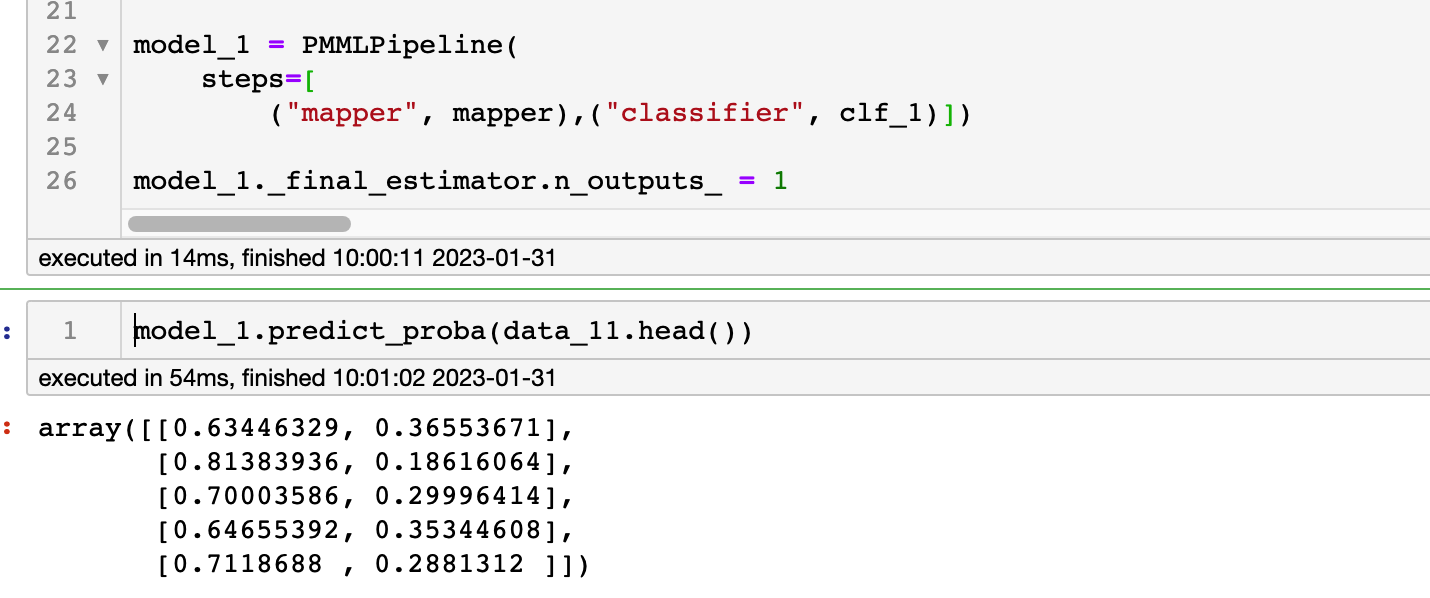
I expect to use the values of certain columns to modify the probability that the prediction is 0, 1. Is it impossible to achieve?