looks like this will be useful.
Closed github-learning-lab[bot] closed 4 years ago
looks like this will be useful.
If you've written your own functions or scripts before, you may have run into the red breakpoint dot :red_circle: on the left side oyour script window:
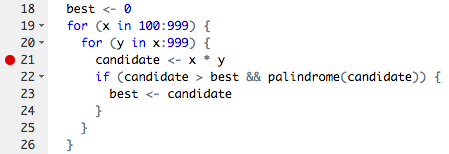
Breakpoints allow you to run a function (or script) up until the line of the breakpoint, and then the evaluation pauses. You are able to inspect all variables available at that point in the evaluation, and even step carefully forward one line at a time. It is out of scope of this exercise to go through exactly how to use debuggers, but they are powerful and helpful tools. It would be a good idea to read up on them if you haven't run into breakpoints yet.
In scipiper, you can't set a breakpoint in the "normal" way, which would be clicking on the line number after you sourced the script. Instead, you need to use the other method for debugging in R, which requires adding the function call browser() to the line where you'd like the function call to stop.
You have a working, albeit brittle, pipeline in your course repository. You can try it out with scipiper::scmake(). This pipeline has a number of things you'll work to fix later, but for now, it is a useful reference. The pipeline contains several functions which are defined in .R files.
So, if you wanted to look at what download_files where created within the download_nwis_data function, you could set a breakpoint by adding browser() to the "1_fetch/src/get_nwis_data.R" file:

Then, running scmake() will land you right in the middle of line 8. Give it a try on your own.
:keyboard: comment on where you think you might find browser() handy in future pipelines.
when something goes wrong also - there's a typo "...if you wanted to look at what `download_files were created..."
Seeing the structure of a pipeline as a visual is powerful. Viewing connections between targets and the direction data is flowing in can help you better understand the role of pipelines in data science work. Once you are more familiar with pipelines, using the same visuals can help you diagnose problems.
Below is a remakefile that is very similar to the one you have in your code repository (the packages and sources fields were removed for brevity, but they are unchanged):
targets:
all:
depends: 3_visualize/out/figure_1.png
site_data:
command: download_nwis_data()
1_fetch/out/site_info.csv:
command: nwis_site_info(fileout = '1_fetch/out/site_info.csv', site_data)
1_fetch/out/nwis_01427207_data.csv:
command: download_nwis_site_data('1_fetch/out/nwis_01427207_data.csv')
1_fetch/out/nwis_01435000_data.csv:
command: download_nwis_site_data('1_fetch/out/nwis_01435000_data.csv')
site_data_clean:
command: process_data(site_data)
site_data_annotated:
command: annotate_data(site_data_clean, site_filename = '1_fetch/out/site_info.csv')
site_data_styled:
command: style_data(site_data_annotated)
3_visualize/out/figure_1.png:
command: plot_nwis_timeseries(fileout = '3_visualize/out/figure_1.png',
site_data_styled, width = 12, height = 7, units = I('in'))Two file targets ("1_fetch/out/nwis_01427207_data.csv" and "1_fetch/out/nwis_01435000_data.csv") were added to this remakefile, but there were no changes to the functions, since download_nwis_site_data() already exists and is used to create a single file that contains water monitoring information for a single site.
The remake package has a nice function called diagram() that we haven't covered yet. It produces a dependency diagram for the target(s) you specify (remember that all is the default target). For this modified remakefile, calling that function with the default arguments produces:
remake::diagram()
If you run the same command, you'll see something similar but the two new files won't be included.
Seeing this diagram helps develop a greater understanding of some of the earlier concepts from intro-to-pipelines. Here, you can clearly see the connection between all and "3_visualize/out/figure_1.png". The figure_1 plot needs to be created in order to complete all. The arrows communicate the connections (or "dependencies") between targets, and if a target doesn't have any arrows connected to it, it isn't depended on by another target and it doesn't depend on any another targets. The two new .csv files are both examples of this, and in the image above they are floating around with no connections. A floater target like these two won't be built by scmake() unless it is called directly (e.g., scmake("1_fetch/out/nwis_01427207_data.csv"); remember again "skip the work you don't need").
The diagram also shows how the inputs of one function create connections to the output of that function. site_data is used to build site_data_clean (and is the only input to that function) and it is also used as an input to "1_fetch/out/site_info.csv", since the nwis_site_info() function needs to know what sites to get information from. These relationships result in a split in the dependency diagram where site_data is directly depended on by two other targets.
By modifying the recipe for the all target, it is possible to create a dependency link to one of the .csv files, which would then result in that file being included in a build as it becomes necessary in order to complete all:
targets:
all:
depends: ["3_visualize/out/figure_1.png", "1_fetch/out/nwis_01427207_data.csv"]And after calling remake::diagram(), it is clear that now "1_fetch/out/nwis_01427207_data.csv" has found a home and is relevant to all!

With this update, the build of "1_fetch/out/nwis_01427207_data.csv" would no longer be skipped when scmake() is called.
:keyboard: comment on what you learned from exploring remake::diagram()
I had to install some new packages for remake::diagram() to work. It also became clear that not all the targets of remake are files. You can have R objects as targets and pass those as inputs into other steps in the workflow. In all the scipiper pipelines that I've seen thus far, I've only seen files on disk as targets.
In the image contained within the previous comment, you may have noticed the faded fill color of the target shapes. That styling signifies that the targets are out of date ("dirty") or haven't been created yet.
We've put some fragile elements in the pipeline that will be addressed later, but if you were able to muscle through the failures with multiple calls to scmake(), you likely were able to build the figure near the end of the dependency diagram. For this example, we'll stop short of building the "3_visualize/out/figure_1.png" target by calling scmake('site_data_styled') instead to illustrate the concept of a dirty target.
The updated remake::diagram() output looks like this:

Only the colors have changed from the last example, signifying that the darker targets are "complete", but that "3_visualize/out/figure_1.png" and the two data.csv files still don't exist.
the scipiper package has a useful function called which_dirty() which will list the incomplete ("dirty") targets that need to be updated in order to satisfy the output (once again, the default for this function is to reference the all target).
which_dirty()
[1] "1_fetch/out/nwis_01427207_data.csv" "3_visualize/out/figure_1.png" "all" This output tells us the same thing as the visual, namely that these three targets :point_up: are incomplete/dirty. No information is shared for the "1_fetch/out/nwis_01435000_data.csv" target, even though it is out of date (and would be considered "dirty" as well). This is because that target is not relevant to building all.
Calling why_dirty() on a single target tells us a number of important things
why_dirty("3_visualize/out/figure_1.png")
The target '3_visualize/out/figure_1.png' does not exist
# A tibble: 4 x 8
type name hash_old hash_new hash_mismatch dirty dirty_by_descent current
<chr> <chr> <chr> <chr> <lgl> <lgl> <lgl> <lgl>
1 target 3_visualize/out/figure_1.png none none FALSE TRUE FALSE FALSE
2 depends site_data_styled NA 183e7990d33bbc76314aa48f04e58531 NA FALSE FALSE TRUE
3 fixed NA NA 96653adafc4622c2088c81ea947966af NA FALSE FALSE TRUE
4 function plot_nwis_timeseries NA 4592eea358bfd73d90bee824dda0e0c7 NA FALSE FALSE TRUE From this output, with a little help from the ?scipiper::why_dirty() documentation, it is clear that "3_visualize/out/figure_1.png" is "dirty" for several reasons:
hash_old and hash_new are both "none"; the function also printed a message _The target '3_visualize/out/figure1.png' does not existsite_data_styled target, the plot_nwis_timeseries function, and the "fixed" inputs to the function (which for this target, include width = 12, height = 7, and units = 'in')A build of the figure with scipiper::scmake('3_visualize/out/figure_1.png') will update the target dependencies, result in a remake::diagram() output which darkens the fill color on the "3_visualize/out/figure_1.png", and cause a call to why_dirty("3_visualize/out/figure_1.png") to result in a descriptive error letting the user know the target is not dirty.
The target will be out of date if there are any modifications to the upstream dependencies (follow the arrows in the diagram "upstream") or to the function plot_nwis_timeseries(). Additionally, a simple update to the value of one of the "fixed" arguments will cause the "3_visualize/out/figure_1.png" target to be "dirty". Here the height argument was change from 7 to 8)
why_dirty("3_visualize/out/figure_1.png")
Since the last build of the target '3_visualize/out/figure_1.png':
* the fixed arguments (character, logical, or numeric) to the target's command have changed
# A tibble: 4 x 8
type name hash_old hash_new hash_mismatch dirty dirty_by_descent current
<chr> <chr> <chr> <chr> <lgl> <lgl> <lgl> <lgl>
1 target 3_visualize/out/figure_1.png bfdee70b50f05636b06ad32ef3b11810 bfdee70b50f05636b06ad32ef3b11810 FALSE TRUE FALSE FALSE
2 depends site_data_styled 183e7990d33bbc76314aa48f04e58531 183e7990d33bbc76314aa48f04e58531 FALSE FALSE FALSE TRUE
3 fixed NA 96653adafc4622c2088c81ea947966af 82eb1fa6b001fe33b8af3c8a629421a8 TRUE FALSE FALSE TRUE
4 function plot_nwis_timeseries 4592eea358bfd73d90bee824dda0e0c7 4592eea358bfd73d90bee824dda0e0c7 FALSE FALSE FALSE TRUE The "hash" of these three fixed input arguments was "96653adafc4622c2088c81ea947966af" and is now "82eb1fa6b001fe33b8af3c8a629421a8", resulting in a "hash_mismatch" and causing "3_visualize/out/figure_1.png" to be "dirty". You'll hear more about hashes in the future, but for now, think of a hash as a string that is unique for any unique data. In the case of fixed arguments, changing the argument names, values, or even the order they are specified will create a different hash value and cause the output target to be considered dirty.
:keyboard: using which_dirty() and why_dirty() can reveal unexpected connections between the target and the various dependencies. Comment on some of the different information you'd get from why_dirty() that wouldn't be available in the visual produced with remake::diagram().
The diagram just shows the dependency graph and whether a target is up to date. why_dirty helps you understand why a given target is not up to date.
using remake::diagram() shows the dependency diagram of the pipeline. Look at previous comments to remind yourself of these visuals.
As a reminder, the direction of the arrows capture the dependency flow, and site_data sits at the top, since it is the first target that needs to be built.
Also note that there are no backward looking arrows. What if site_data relied on site_data_styled? In order to satisfy that relationship, an arrow would need to swing back up from site_data_styled and connect with site_data. Unfortunately, this creates a cyclical dependency since changes to one target change the other target and changes to that target feed back and change the original target...so on, and so forth...
This potentially infinite loop is confusing to think about and is also something that dependency managers can't support. We won't say much more about this issue here, but note that in the early days of building pipelines if you run into a cyclical dependency error, this is what's going on.
:keyboard: Add a comment when you are ready to move on.
gotcha
Moving into a pipeline-way-of-thinking can reveal some suprising habits you created when working under a different paradigm. Moving the work of scripts into functions is one thing that helps compartmentalize thinking and organize data and code relationships, but smart pipelines require even more special attention to how functions are designed.
It is tempting to build functions that do several things; perhaps a plotting function also writes a table, or a data munging function returns a data.frame, but also writes a log file. You may have noticed that there is no easy way to specify two ouputs from a single function in a pipeline recipe. We can have multiple files as inputs into a function/command, but only one output/target. If a function writes a file that is not explicitly connected in the recipe (i.e., it is a "side-effect" output), the file is untracked by the dependency manager, and treated like an implicit file target (i.e., a target which has no "command" to specify how it is built; future issues will cover more details on this type of target). If the side-effect file is relied upon by a later target, changes to the side-effect target will indeed trigger a rebuild of the downstream target, but the dependency manager will have no way of knowing when the side-effect target itself should be rebuilt. :no_mobile_phones:
Maybe the above doesn't sound like a real issue, since the side-effect target would be updated every time the other explicit target it is paired with is rebuilt. But this becomes a scary problem (and our first real gotcha!) if the explicit target is not connected to the critical path of the final sets of targets you want to build, but the implicit side-effect target is. What this means is that even if the explicit target is out of date, it will not be rebuilt because building this target is unnecessary to completing the final targets (remember "skip the work you don't need" :arrow_right_hook:). The dependency manager doesn't know that there is a hidden rule for updating the side-effect target and that this update is necessary for assuring the final targets are up-to-date and correct. :twisted_rightwards_arrows:
Side-effect targets can be used effectively, but doing so requires a good understanding of implications for tracking them and advanced strategies on how to specify rules and dependencies in a way that carries them along. :ballot_box_with_check:
Additionally, it is tempting to code a filepath within a function which has information that needs to be accessed in order to run. This seems harmless, since functions are tracked by the dependency manager and any changes to those will trigger rebuilds, right? Not quite. If a file like "1_fetch/in/my_metadata.csv" is specified in a function, but is not also an argument to the command in the remakefile recipe or listed as a depends of the resulting target, any changes to the "1_fetch/in/my_metadata.csv" will go unnoticed by the dependency manager, since the string that specifies the file name remains unchanged. The system isn't smart enough to know that it needs to check whether that file has changed.
As a rule, unless you are purposefully trying to hide changes in a file from the dependency manager, do not read non-argument files in the body of a function. :end:
:keyboard: Add a comment when you are ready to move on
10-4
You might have a project where there is a directory :file_folder: with a collection of files. To simplify the example, assume all of the files are .csv and have the same format. As part of the hypothetical project goals, these files need to be combined and formatted into a single plottable data.frame.
In a data pipeline, we'd want assurance that any time the number of files changes, we'd rebuild the resulting data.frame. Likewise, if at any point the contents of any one of the files changes, we'd also want to re-build the data.frame.
This hypothetical example could be coded as
sources:
- combine_files.R
targets:
all:
depends: figure_1.png
plot_data:
command: combine_into_df('1_fetch/in/file1.csv','1_fetch/in/file2.csv','1_fetch/in/file3.csv')
figure_1.png:
command: my_plot(plot_data):point_up: This coding would work, as it tells the dependency manager which files to track for changes, and where the files in the directory are. But this solution is less than ideal, both because it doesn't scale well to many files, and because it doesn't adapt to new files coming into the 1_fetch/in directory :file_folder: (the pipeline coder needs to manually add file names)
Alternatively, what about adding the directory as an input to the recipe, like this (you'd also need to modify your combine_into_df function to use dir(work_dir) to generate the file names)?
sources:
- combine_files.R
targets:
all:
depends: figure_1.png
plot_data:
command: combine_into_df(work_dir = '1_fetch/in')
figure_1.png:
command: my_plot(plot_data)After running scmake(), :point_up: this looks like it works, but results in a rather cryptic warning message might end up being ignored:
Warning messages:
1: In structure(.Call(C_Rmd5, files), names = files) :
md5 failed on file '1_fetch/in'An explaination of that warning message - In order to determine if file contents have changed, remake uses md5sum() from the tools package to create a unique hash for each target. You can take a look at what this does:
tools::md5sum('3_visualize/out/figure_1.png')
3_visualize/out/figure_1.png
"500a71be79d1e45457cdb2c31a03be46" md5sum() fails when you point it toward a directory, since it doesn't know what to do with that kind of input.
A third strategy might be to create a target that lists the contents of the directory, and then uses that list as an input:
sources:
- combine_files.R
targets:
all:
depends: figure_1.png
work_files:
command: dir('1_fetch/in')
plot_data:
command: combine_into_df(work_files)
figure_1.png:
command: my_plot(plot_data):point_up: This approach is close, but has a few flaws: 1) because '1_fetch/in' can't be tracked as a real target, changes within that directory aren't tracked (same issue as the previous example), and 2) if the contents of the files change, but the number and names of the files don't change, the work_files target won't appear changed, and therefore the necessary rebuild of plot_data would be skipped (another example of "skip the work you don't need" in action)
Instead, we would avocate for a modified approach that combines the work of md5sum with the collection of files returned with dir. If instead of work_files being a vector of file names (in this case, c('1_fetch/in/file1.csv','1_fetch/in/file2.csv','1_fetch/in/file3.csv')) - can we make it a data.frame of file names and their associated hashes?
# A tibble: 3 x 2
filepath hash
<chr> <chr>
1 1_fetch/in/file1.csv 530886e2b604d6d32df9ce2785c39672
2 1_fetch/in/file2.csv dc36b6dea28abc177b719015a18bccde
3 1_fetch/in/file3.csv c04f87fb9af74c4e2e2e77eed4ec80f3Now, if the combine_into_df can by modified to accept this type of input (i.e., simply grab the filepath with $filepath), then we'll have an input that both reflects the total number of files in the directory, and captures a reference hash that will reveal any future changes. Yay! :star2: This works because a change to any one of those hashes will result in a change in the hash of the new modified work_files data.frame, which then would cause a rebuild of plot_data.
But unfortunately, because work_files still relies on a directory and therefore can't be tracked, we're still missing a critical part of the solution. In the next topic, we'll reveal a more elegant way to trigger rebuilds, but for now, note you'd need to manually run
scmake('work_files', force = TRUE) to force scipiper to dig into that directory and hash all of the files. If the result of work_files that comes back is the same as before, running scmake() won't build anything new and we can be confident that figure_1.png is up to date.
:keyboard: Add a comment when you are ready to move on.
huh. cool. I'd never thought about doing something like that.
also there's a typo @jread-usgs/@aapling-usgs - "Now, if the combine_into_df can be ..."
Wow, we've gotten this far and haven't written a function that accepts anything other than an object target or a file target. I feel so constrained!
In reality, R functions have all kinds of other arguments, from logicals (TRUE/FALSE), to characters that specify which configurations to use.
The example in your working pipeline creates a figure, called 3_visualize/out/figure_1.png. Unless you've made a lot of modifications to the plot_nwis_timeseries() function, it has a few arguments that have default values, namely width = 12, height = 7, and units = 'in'. Nice, you can control your output plot size here!
But adding those to the remake file like so
3_visualize/out/figure_1.png:
command: plot_nwis_timeseries(target_name, site_data_styled, width = 12, height = 7, units = 'in')causes an immediate error
scmake()
Error in .remake_add_targets_implied(obj) :
Implicitly created targets must all be files:
- in: (in 3_visualize/out/figure_1.png) -- did you mean: site_data, 1_fetch/out/site_info.csv, Since 'in' is not an object target with a recipe in the pipeline, remake is trying to find the file corresponding to "in", since it must be a file if it isn't a number or an object.
We know "in" is not a file, it is instead a simple argument we want to expose in the recipe, so we can make modification. To do this, wrapping the argument in the I() function tells remake to "treat this argument 'as is'", meaning don't try to infer anything fancy, just pass it along to the function.
3_visualize/out/figure_1.png:
command: plot_nwis_timeseries(target_name, site_data_styled, width = 12, height = 7, units = I('in'))works! :star2:
Going back to our previous example where we wanted to build a data.frame of file hashes from the '1_fetch/in' directory, I() comes in handy in two spots: 1) we can avoid the md5sum warning by wrapping the directory path in the I(), which means '1_fetch/in' is used as a real string instead of attempting to treat it as a file target, and 2) we can add a new variable that can help us force or refresh the target with some information about when we last did it.
sources:
- combine_files.R
targets:
all:
depends: figure_1.png
work_files:
command: hash_dir_files(directory = I('1_fetch/in'), dummy = I('2020-05-18'))By adding this dummy argument to our own custom hash_dir_files() function, we can modify the dummy contents any time we want to force the update of work_files. Making a change to the text in the dummy argument has the same outcome as scmake('work_files', force = TRUE), but this way we keep an easy to read record of when we last manually refreshed the pipeline's view of what exists within '1_fetch/in'. You may see the use of these dummy arguments in spots where there is no other trigger that would cause a rebuild (such as the case of these directory challenges or when pulling data from a remote webservice or website - scipiper has no way of knowing that new data are available on the same service URL).
:keyboard: Add a comment when you are ready to move on.
Clever. Good to know. So we use I to tell scipiper not to try to interpret that argument, but rather just pass it along.
target_name special variable.target:command relationships and reducing duplicationIn your repo, there is a remake files that specifies relationships between targets, commands, and other targets.
For one of the targets, you'll see this:
1_fetch/out/site_info.csv:
command: nwis_site_info(fileout = '1_fetch/out/site_info.csv', site_data)It seems odd to duplicate the name of the target twice. First it is used to declare the file target path as 1_fetch/out/site_info.csv. But then it is used a second time because the nwis_site_info needs to know the name of the file to write the downloaded data too. Doesn't this seem duplicative and potentially dangerous (what if you make a typo to the fileout argument and it writes to a different file)?
The target_name variable is a useful way to avoid the duplication and danger of repeating the name of the target as one of the inputs. We can instead write the target recipe as
1_fetch/out/site_info.csv:
command: nwis_site_info(fileout = target_name, site_data)Here, target_name will tell scipiper to use "1_fetch/out/site_info.csv" for the fileout argument in the nwis_site_info function when building the target. Using target_name not only saves us some time and makes the construction of pipelines less error prone, it also helps us create patterns that can be used to generate multiple targets in a simpler fashion.
Imagine part of the target name is used to specify something within the function call, such as a site number.
1_fetch/out/nwis_01427207_data.csv:
command: download_nwis_site_data(target_name)
1_fetch/out/nwis_01432160_data.csv:
command: download_nwis_site_data(target_name)
1_fetch/out/nwis_01436690_data.csv:
command: download_nwis_site_data(target_name)
the download_nwis_site_data function uses a regular expression to extract the site number from the input file name in order to make a web service request for data. Here, even though it seems goofy to create so many download targets, we reduce the places mistakes can be made by using target_name instead of repeating the use of the file name.
:keyboard: Add a comment when you are ready to move on.
That's handy
In this section, we're going to go one by one through a series of tips that will help you avoid common pitfalls (or gotchas!) in pipelines. These tips will help you in the next sections and in future work. A quick list of what's to come:
which_dirty()andwhy_dirty()to further interrogate the status of pipeline targetsI()helpertarget_namespecial variable. Simplifyingtarget:left_right_arrow:commandrelationships and reducing duplication:keyboard: add a comment to this issue and the bot will respond with the next topic
I'll sit patiently until you comment