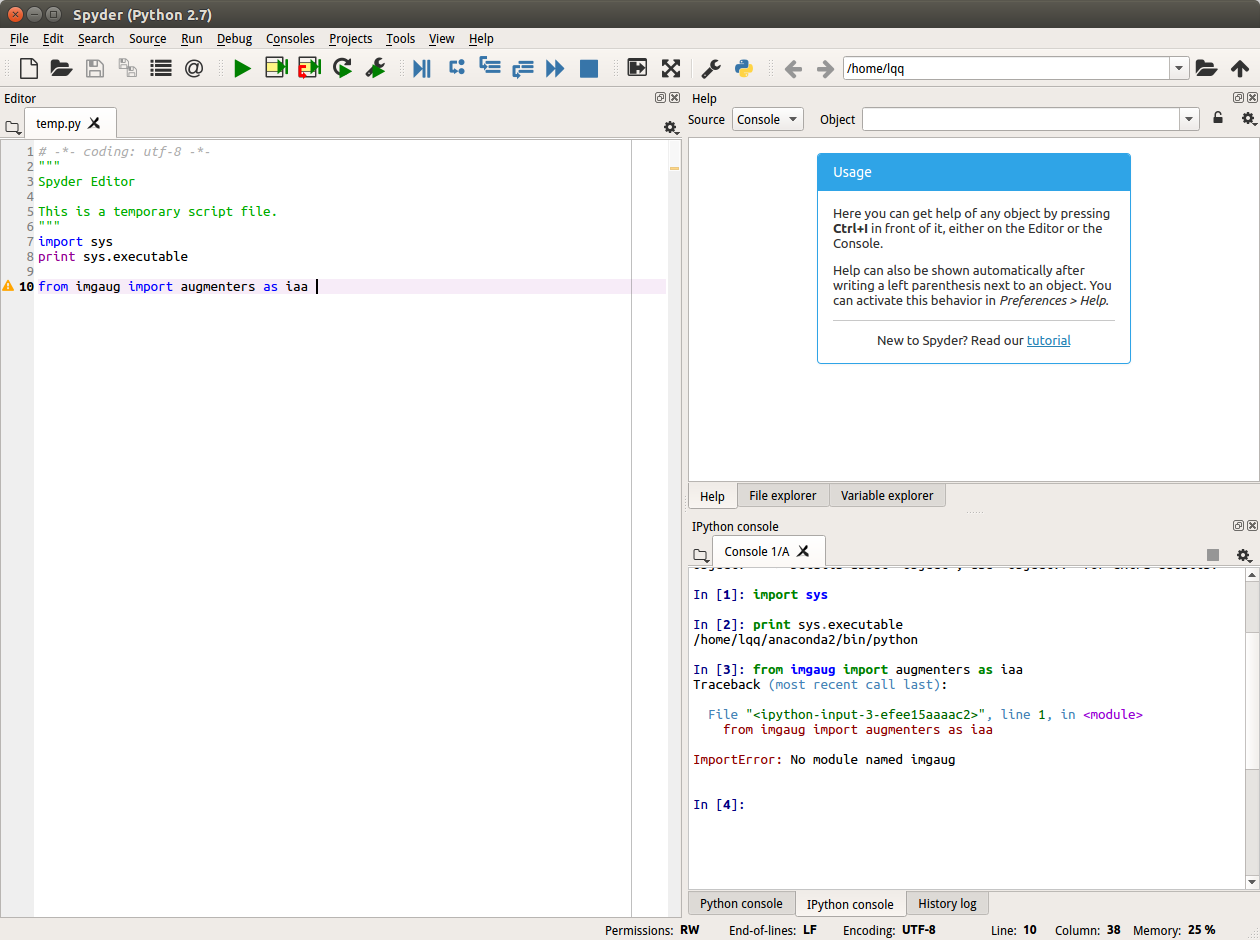
Usually that indicates that the notebook is running with a different Python or in a different environment from Python in the command prompt. Check sys.executable to see which Python it's running in, and sys.path to see where it's looking for imports.






Hi,
I am new to Python and Anaconda. I installed anaconda and install Scipy. When I try import scipy in the Python in command prompt on the Anaconda prompt, it works fine as below
But when I use the same in the Jupyter notebook, I am getting -
Please let me know how to solve this issue.