The layout algorithm is not perfect and there may be rare corner cases where it fails to layout the regions. You should be able to see how the circles should look like using the venn3_circles method (which only plots the circles, without going through the steps of splitting them into regions).
It would be nice if you could provide the actual subset sizes which lead to this error. Try calling
matplotlib_venn._venn3.compute_venn3_subsets(a,b,c)(where a,b,c are your three sets) and posting the resulting 7-number tuple here.


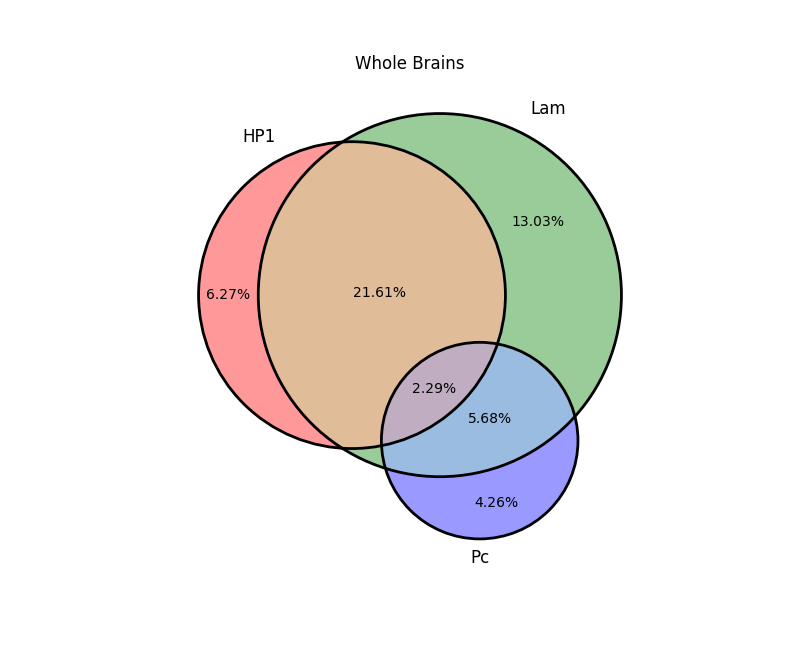 What 'hack' can I use to get rid of little green triangle in the bottom?
The numbers were: 7549417, 15685620, 26018311, 5128906, 301048, 6841264, 2762301
What 'hack' can I use to get rid of little green triangle in the bottom?
The numbers were: 7549417, 15685620, 26018311, 5128906, 301048, 6841264, 2762301
Hi!
I'm using venn3 to plot 3 sets of names and the following exception appeared:
I can't figure out what is exactly going on and whether there is something I can do to solve it. I bit of help would be appreciated. Thanks!