Codecov Report
Merging #44 into master will increase coverage by
0.77%. The diff coverage isn/a.
@@ Coverage Diff @@
## master #44 +/- ##
==========================================
+ Coverage 68.5% 69.28% +0.77%
==========================================
Files 17 17
Lines 1810 1810
Branches 166 166
==========================================
+ Hits 1240 1254 +14
+ Misses 538 527 -11
+ Partials 32 29 -3| Impacted Files | Coverage Δ | |
|---|---|---|
| PyEMD/CEEMDAN.py | 94.26% <ø> (+4.91%) |
:arrow_up: |
| PyEMD/EMD.py | 87.5% <0%> (+0.24%) |
:arrow_up: |
| PyEMD/tests/test_eemd.py | 100% <0%> (+3.17%) |
:arrow_up: |
| PyEMD/EEMD.py | 83.58% <0%> (+7.46%) |
:arrow_up: |
Continue to review full report at Codecov.
Legend - Click here to learn more
Δ = absolute <relative> (impact),ø = not affected,? = missing dataPowered by Codecov. Last update de373ec...2837477. Read the comment docs.
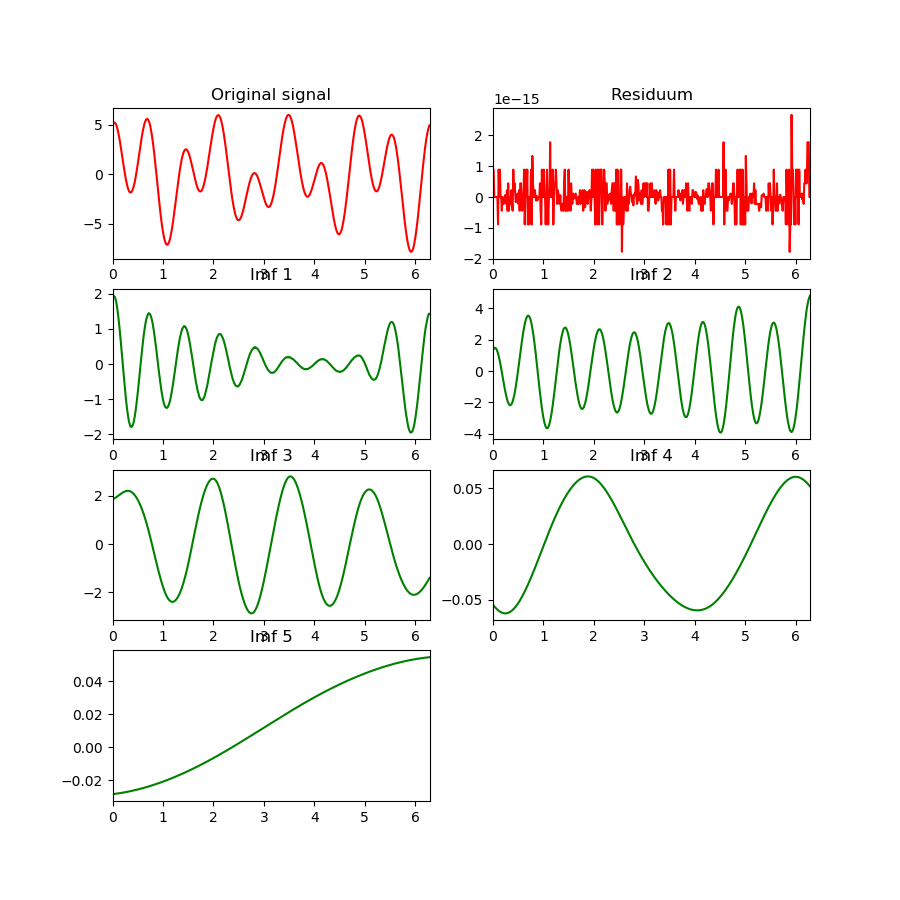
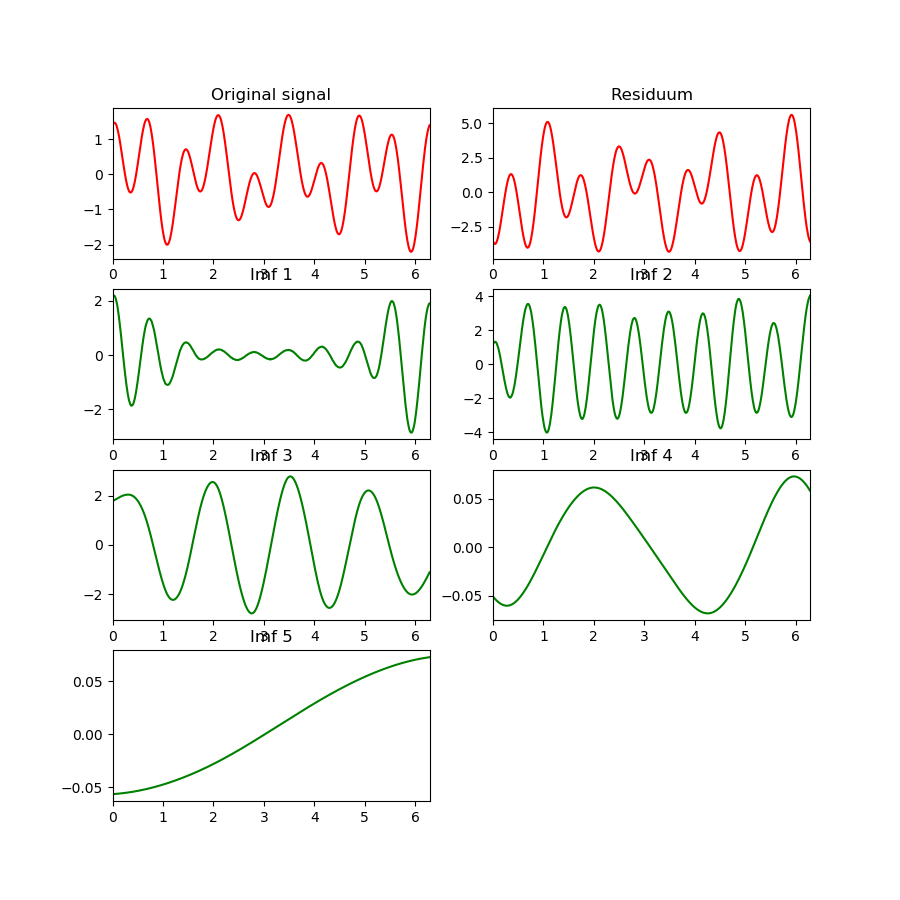 After:
After:
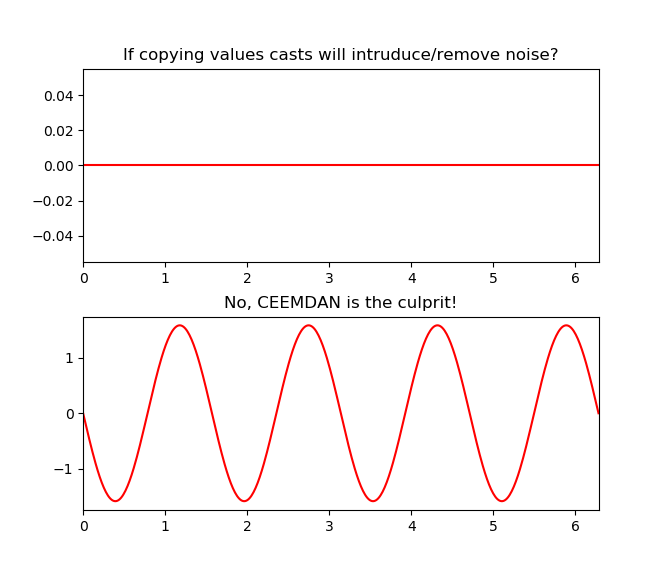 You can find the value of
You can find the value of
42