Please copy your readme info here
Open veritas496 opened 2 years ago
Please copy your readme info here
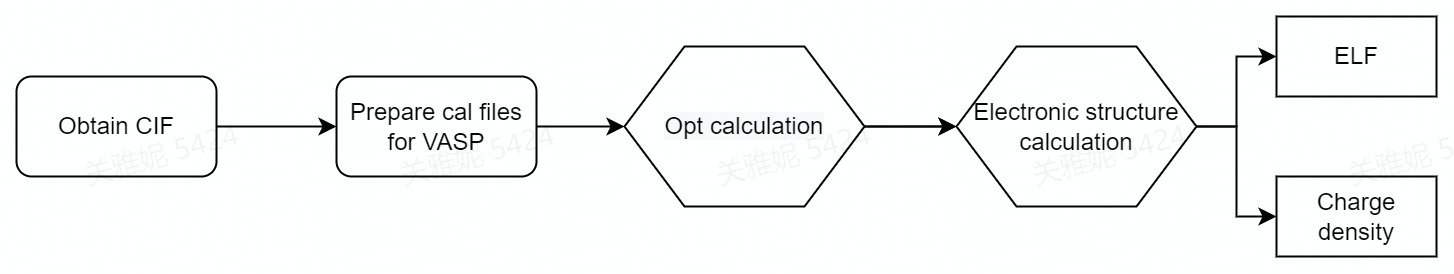
This dflow proposal is for electronic structure calculations using VASP, from obtaining cif structure information to structure optimization, and then to calculate several electronic structure properties in parallel, ELF and charge density as examples in this project.
Traditionally, we need to obtain the .cif file containing structure information from some database, e.g. Material Project, and then prepare calculation files required for VASP calculation. After that, it is the time to submit a structure optimization task and then with optimized structure, we need to modify parameters in INCAR and then submit a calculation task for ELF, and then modify parameters in INCAR to submit another calculation task for charge density.
However, with the aid of dflow, the workflow can be achieve automatically. The dflow workflow diagram is shown in Figure 1.
input:
output:
execute:
class DownloadCIF(OP):
def __init__(self):
pass
@classmethod
def get_input_sign(cls):
return OPIOSign({
'api-key' : str,
'mp-id' : int,
})
@classmethod
def get_output_sign(cls):
return OPIOSign({
'DownloadCIF_output' : Artifact(Path),
})
@OP.exec_sign_check
def execute(self, op_in : OPIO):
from pymatgen.ext.matproj import MPRester
from pymatgen.io.cif import CifWriter
api_key=op_in['api-key']
mp_id=str(op_in['mp-id'])
mpr=MPRester(str(api_key))
structure=mpr.get_structure_by_material_id(f'mp-{mp_id}',final=True,conventional_unit_cell=True)
CifWriter(structure).write_file(f'{mp_id}.cif')
return OPIO({
"DownloadCIF_output": Path(f'{mp_id}.cif')
})class VASPPrep(OP):
def __init__(self):
pass
@classmethod
def get_input_sign(cls):
return OPIOSign({
'VaspPre_input': Artifact(Path)
})
@classmethod
def get_output_sign(cls):
return OPIOSign({
'VaspPre_output': Artifact(Path)
})
@OP.exec_sign_check
def execute(self, op_in: OPIO) -> OPIO:
from pymatgen.core import Structure
from pymatgen.io.vasp.sets import MPRelaxSet, MITRelaxSet
structureOpt = Structure.from_file(filename=op_in['VaspPre_input'])
relax_set = MPRelaxSet(structure=structureOpt)
# relax_set = MITRelaxSet(structure=optStructure)#MPRelaxSet(structure=optStructure)
relax_set.write_input(output_dir="VaspOpt")
return OPIO({"VaspPre_output" : Path('./VaspOpt')})input:
execute:
class VASPOpt(OP):
def __init__(self):
pass
@classmethod
def get_input_sign(cls):
return OPIOSign({
"VaspOpt_input": Artifact(Path),
"running_cores": int
})
@classmethod
def get_output_sign(cls):
return OPIOSign({
"VaspOpt_output": Artifact(Path)
})
@OP.exec_sign_check
def execute(self, op_in: OPIO) -> OPIO:
cwd = os.getcwd()
os.chdir(op_in["VaspOpt_input"])
cmd = f'source /public1/soft/intel/2020/parallel_studio_xe_2020/psxevars.sh &&\
mpirun -np {str(op_in["running_cores"])} /your-VASP-execute-environment/bin/vasp_vtst_std'
subprocess.call(cmd, shell=True)
os.chdir(cwd)
return OPIO({
"VaspOpt_output": Path(op_in["VaspOpt_input"])/"CONTCAR",
})class VASPSingle(OP):
def __init__(self):
pass
@classmethod
def get_input_sign(cls):
return OPIOSign({
"VaspSingle_input_optModel": Artifact(Path),
"VaspSingle_input": Artifact(Path),
"running_cores": int,
"INCARfromOpt": dict,
"AIMCAR": str
})
@classmethod
def get_output_sign(cls):
return OPIOSign({
"VaspSingle_output": Artifact(Path)
})
@OP.exec_sign_check
def execute(self, op_in: OPIO) -> OPIO:
cwd = os.getcwd()
os.system(f'mv {op_in["VaspSingle_input_optModel"]}/CONTCAR {op_in["VaspSingle_input"]}/POSCAR')
os.chdir(op_in["VaspSingle_input"])
from pymatgen.io.vasp import inputs
incar_opt = inputs.Incar().from_file("INCAR")
for key, value in op_in["INCARfromOpt"].items():
incar_opt[key] = value
incar_opt.write_file("INCAR")
cmd = f'source /public1/soft/intel/2020/parallel_studio_xe_2020/psxevars.sh &&\
mpirun -np {str(op_in["running_cores"])} /your-VASP-execute-environment/bin/vasp_vtst_std'
subprocess.call(cmd, shell=True)
os.chdir(cwd)
return OPIO({
"VaspSingle_output": Path(op_in["VaspSingle_input"])/op_in["AIMCAR"],
})You are expected to enter your slurm information in this part, e.g. host, port, username, password, and header.
def slurm_exe(cores: int):
"""
you are expected to enter your slurm information in this part, e.g. host, port, username, password, and header.
"""
slurm_remote_executor = SlurmRemoteExecutor(
host="your host",
port=22,
username="your username",
password="your password",
header=f"#!/bin/bash\n#SBATCH --nodes=1\n#SBATCH --ntasks-per-node={str(cores)}\n#SBATCH --partition=xxx\n#SBATCH -e output.err",
)
return slurm_remote_executorYou are expected to enter your Material Project API key and targeted model's ID. For example, mp-id=30 is for Cu.
def main():
"""
you are expected to enter your Material Project API key and targeted model's ID. For example, mp-id=30 is for Cu.
"""
Download_CIF = Step(
"download-cif",
PythonOPTemplate(DownloadCIF, image="kianpu/pymatgen"),
parameters={"api-key":"your api-key","mp-id":30},
)
VASP_prep = Step(
"vasp-prep",
PythonOPTemplate(VASPPrep,command=["source ~/.start_miniconda.sh && conda activate my_pymatgen && python"]),
artifacts={"VaspPre_input":Download_CIF.outputs.artifacts["DownloadCIF_output"]},
executor=slurm_exe(1),
)
Structure_Opt = Step(
"structure-opt",
PythonOPTemplate(VASPOpt,command=["source ~/.start_miniconda.sh && conda activate my_pymatgen && python"]),
artifacts={"VaspOpt_input": VASP_prep.outputs.artifacts["VaspPre_output"]},
parameters={"running_cores": 64},
executor=slurm_exe(128),
)
Single_elf = Step(
"single-elf",
PythonOPTemplate(VASPSingle, command=["source ~/.start_miniconda.sh && conda activate my_pymatgen && python"]),
artifacts={
"VaspSingle_input_optModel": Structure_Opt.outputs.artifacts["VaspOpt_output"],
"VaspSingle_input": VASP_prep.outputs.artifacts["VaspPre_output"]
},
parameters={
"running_cores":64,
"INCARfromOpt": {"LELF": ".TRUE."},
"AIMCAR": "ELFCAR",
},
executor=slurm_exe(128),
)
Single_density = Step(
"single-density",
PythonOPTemplate(VASPSingle, command=["source ~/.start_miniconda.sh && conda activate my_pymatgen && python"]),
artifacts={
"VaspSingle_input_optModel": Structure_Opt.outputs.artifacts["VaspOpt_output"],
"VaspSingle_input": VASP_prep.outputs.artifacts["VaspPre_output"]
},
parameters={
"running_cores":64,
"INCARfromOpt": {"LCHARG":".TRUE.", "NSW": 0},
"AIMCAR": "CHGCAR",
},
executor=slurm_exe(128),
)
wf = Workflow(name="vasp-task")
wf.add(Download_CIF)
wf.add(VASP_prep)
wf.add(Structure_Opt)
wf.add([Single_elf, Single_density])
wf.submit()
if __name__ == "__main__":
main()ok please make a pr
To Whom It May Concern,
Please find the enclosed link for my dflow proposal.
This project is for electronic structure calculation. Traditionally, this kind of calculation tasks is repeated and requires similar calculation files. In other words, in most case we just need to modify corresponding parameters in INCAR. This structured workflow means it can be achieved automatically. With the aid of dflow, we can use all kinds of python package to finish our task. In this work, pymatgen was used to obtain structure information, write cif file, generate calculation files for opt, read INCAR parameters, modify INCAR parameters, and submit tasks in parallel.
This dflow proposal seems simple but definitely can be used further, especially in high throughput calculation.
Please feel free to give your any suggestion and comments.
Best Regards, Yani PhD student