My guess is that you are screwing up the reshaping and/or sizes at least once, maybe multiple times. Just briefly looking at your code, I can already see that you're creating a DenseCRF of shape predictions.shape[1], predictions.shape[0] in the constructor, but after mAP reshaping the result to predictions.shape[0], predictions.shape[1]), which doesn't make sense. If you look closely at the README or examples, you should see that the shape is always consisent. Maybe it's a typo in the constructor call?
Hi,
Thank you for sharing this code. I have a deep learning classifier that specifies probability values for three classes at each pixel in an image. I would like to run the dense CRF to clean up my predictions. Here is the code that I'm trying to use.
`
Define the CRF (width, height, nlabels)
Predictions is a numpy array from my classifier, for a three class classification problem
d = dcrf.DenseCRF2D(predictions.shape[1], predictions.shape[0], 3)
Unary potentials
U = predictions.reshape((3, -1)) d.setUnaryEnergy(-np.log(U))
Pairwise potentials
d.addPairwiseGaussian(sxy=(3,3), compat=3, kernel=dcrf.DIAG_KERNEL, normalization = dcrf.NORMALIZE_SYMMETRIC)
Inference
Q = d.inference(5)
map = np.argmax(Q, axis=0).reshape((predictions.shape[0], predictions.shape[1]))
proba = np.array(map)
Write to a MAT file
scipy.io.savemat("CRF_Predictions.mat", mdict={'proba': proba})`
Here is my original predictions: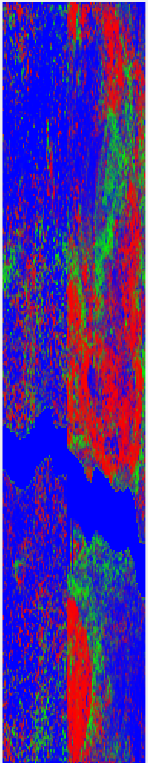
This is the ground truth:
This is what I get after I run the code above: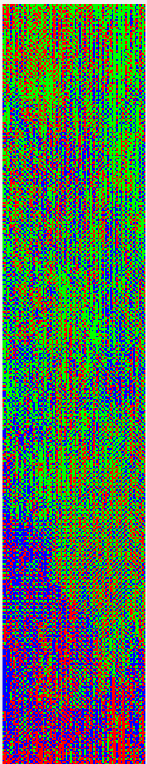
If anyone can tell me where I am making a mistake, that will be really helpful.
Thanks, Chaitanya