Use an arbitrary 3D geometry. Don't let DeepXDE sample the points, but provide the points as anchors in dde.data.PDE.
Open praksharma opened 2 years ago
Use an arbitrary 3D geometry. Don't let DeepXDE sample the points, but provide the points as anchors in dde.data.PDE.
I am solving a 2D steady-state heat conduction problem as follows:
Here, I have parametrized the upper limit of y-boundary condition. For this, I introduced another input in the network i.e.
u(x,y,L).I set the top boundary (the parametrized one) as follows:
The PDE loss reduces the residual as follows:
I set
num_domain = 25000andnum_boundary = 10000.This solution looks good.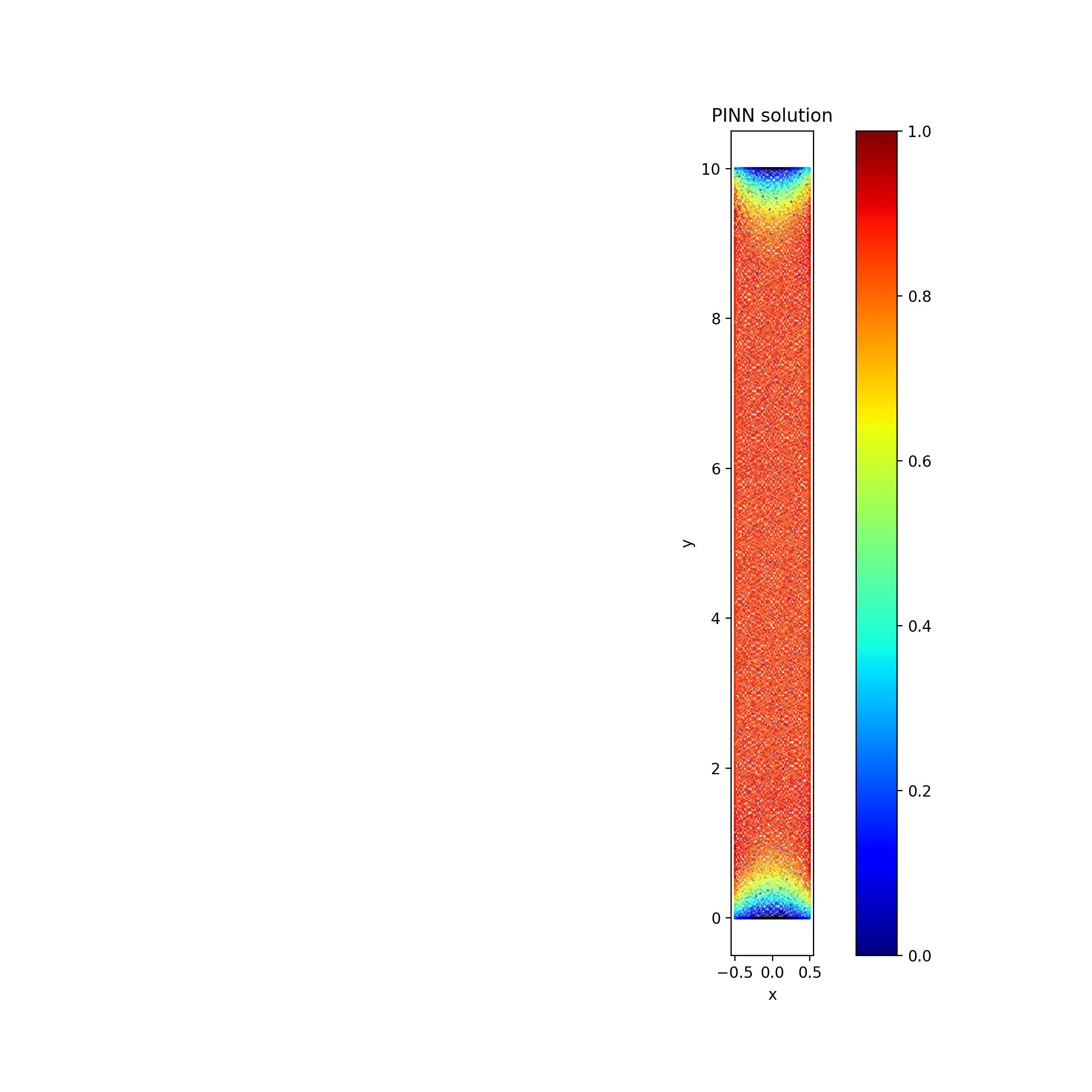
I do not think this is the correct way. The geometry should be variable like this
How to implement a sampling like this in DeepXDE?