This is a start on some basic analyses of nucleotide diversity, I'll try to write up a summary of main points and add in comments below next week.
Closed alimanfoo closed 6 years ago
This is a start on some basic analyses of nucleotide diversity, I'll try to write up a summary of main points and add in comments below next week.
OK, this is ready for some discussion. Here's a summary of main points so far...
Here's a comparison of nucleotide diversity (pi) at the two sampling sites:
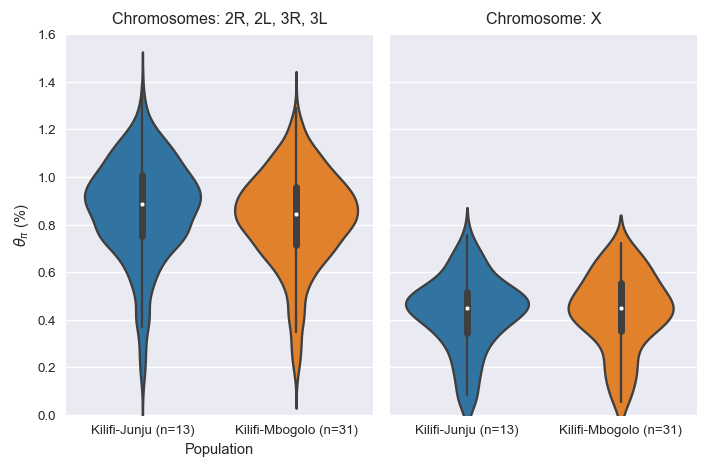
Although it's very similar, in fact there is a small but significantly higher diversity at Junju (pi=0.89%) versus Mbogolo (pi=0.84%):
Nucleotide diversity; chromosomes 2R, 2L, 3R, 3L
------------------------------------------------
Kilifi-Junju (n=13) : median=0.889%; 95% CI [0.876-0.900]
Kilifi-Mbogolo (n=31) : median=0.843%; 95% CI [0.829-0.858]
Kilifi-Junju (n=13) versus Kilifi-Mbogolo (n=31) : Wilcoxon signed rank test P=1.32e-35; statistic=238514.0...although this does not apply to the X chromosome:
Nucleotide diversity; Chromosome X
----------------------------------
Kilifi-Junju (n=13) : median=0.448%; 95% CI [0.429-0.476]
Kilifi-Mbogolo (n=31) : median=0.449%; 95% CI [0.420-0.470]
Kilifi-Junju (n=13) versus Kilifi-Mbogolo (n=31) : Wilcoxon signed rank test P=0.260; statistic=4924.0Here are genome plots comparing diversity at the two sites:
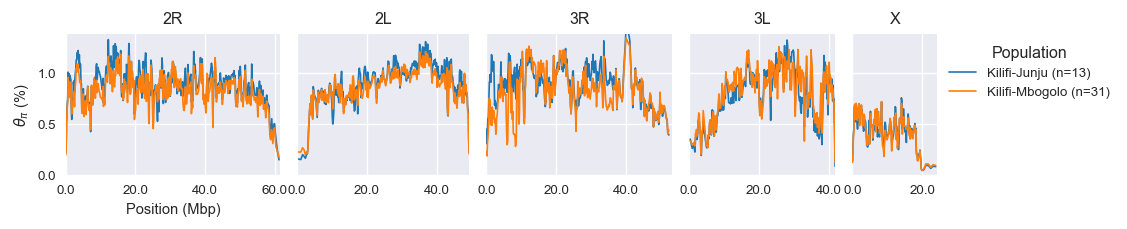
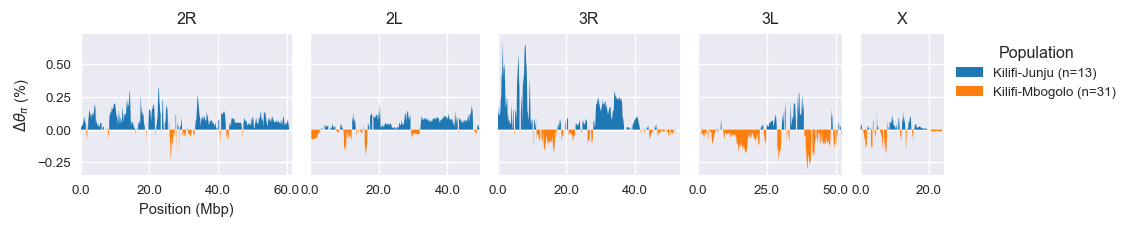
Analysis notebook is here.
cc @jmidega, @hardingnj
I have noted the changes made; Thanks Alistair.
No problem, I'll chat with @hardingnj about where to go next with the analysis.
This PR has some initial work on nucleotide diversity to resolve #8.