I would stay with the vertices. There will be a simple algorithm to select which vertices from resoution n are on the boundary of resolution (n-1) or (n-2) etc. I'll give some thought to this & post another reply :).
There will also be a simple algorithm to determine whether the resulting vertices can be pruned because they form a straight line - essentially iso-latitudinal and iso-longitudinal boundaries are straight in lat, long space, as you note in your side-note, that means only the dart and skew_quad cells need to be processed.
Cell shape can be determined with the function _cell.ellipsoidalshape(self) returns the shape of the cell as one of (‘quad’, ‘cap’, ‘dart’, or ‘skew_quad’) when viewed on the ellipsoid. If you want to do some bulk sorting of which cells need to be called, then in a (lat,long) coordinate space:
- O, P, Q & R faces only have
quadcells, which have four straight boundaries - either iso-latitudnal or iso-longitudinal, - N & S faces all north and south cell boundaries are iso-latitudinal and therefore straight,
- N & S
dartcells andskew-quadcells have a pair of east and west boundaries that need to be interpolated, and - N & S
capcells have no boundaries needing to be interpolated since all their boundaries are either south or north iso-latitudinal boundaries respectively and are therefore all straight.




I'm trying to use the "boundary" method of Cell to get a better outline for dart and skew_quad cells, while displaying a map with cells, cell parents, and parent parents.
I figured I'd need 10 points per side in order for the parents and the "grandsons" to align, which seems to be the case. However, calling boundary is a lot slower than calling vertices, to the point that map display it not interactive any longer.
For reference, this map, produced using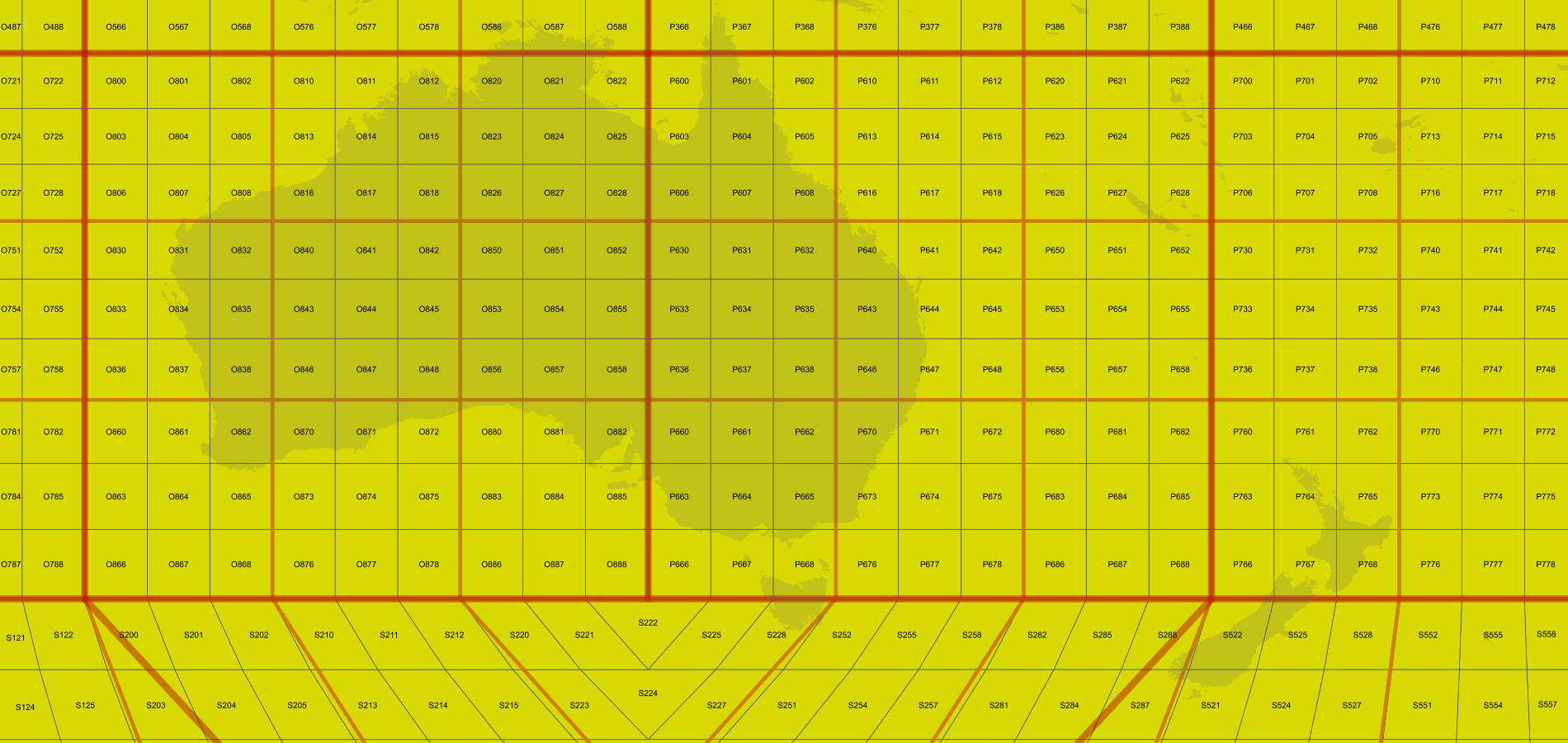
vertices(False), takes 2 seconds, which is slow but still acceptable for an output of this size (click to see full size):This one instead, produced using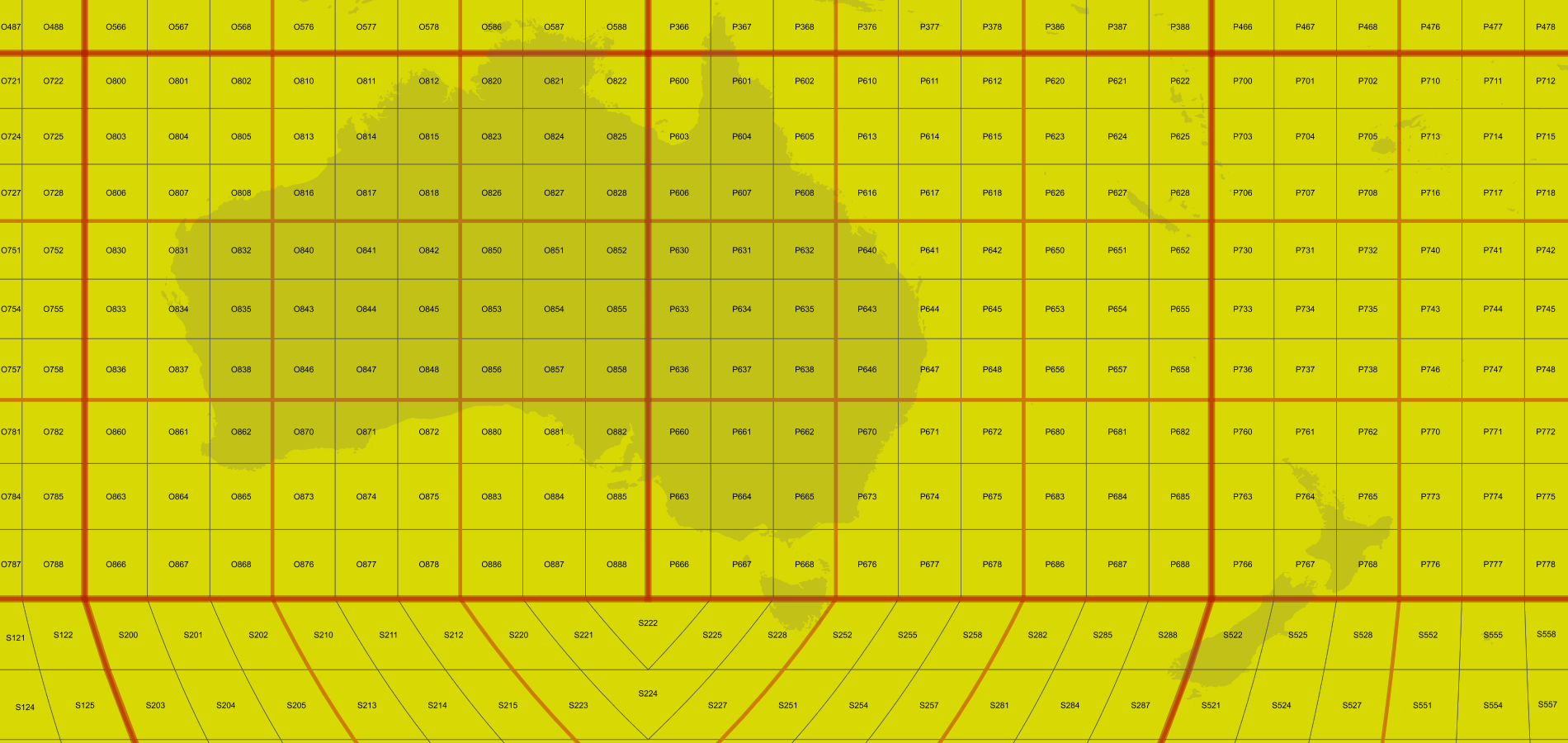
boundary(10, False), requires 14 seconds, which is too much even for a large output:I've reduced it to a pure python script and tested the results with "time", to make sure it was not the GeoServer Java code calling onto the interpreter, slowing down things.
Vertices test
Boundary test
And output:
Boundary does more work, so it's normal that it would be slower... but the overhead seem to be too big.
Side note, it would probably interesting to compromise and call the boundary method only on skew_quad and dart cells, but I don't see a method to determine the type of the cell. Is there any obvious test that can be performed to lean about the nature of the cell? I'm guessing that maybe it's just a matter of checking if the top-most parent is either N or S? (edit, this approach seems to work, reduces the time of the "accurate" map down to 6 seconds... which is still too much).