Sounds nice! Although I don't actually know the breakdown of time spent in the distance calculation versus binning these days. Do you know?
Closed manodeep closed 2 months ago
Sounds nice! Although I don't actually know the breakdown of time spent in the distance calculation versus binning these days. Do you know?
Previously, almost all the time is in the histogram update but that's expected since the histogram update is in the innermost loop.
Hard to say with the new concept - since with ?GEMM, we could compute the separation in (big) chunks and then do the histogram on chunks than the current max chunksize of 16 on fp32 + AVX512. We would also benefit from the other optimisation where the number of bins is 1 for far apart cell-pairs. We would only need to write the FALLBACK version for such a code - the compiler should be able to vectorise it well.
//For the case of a single-histogram bin and no weightavg / rpavg
call_to_gemm() //possibly in chunks, depending on how big the resultant pairwise matrix will be (has to have an user-supplied buffer matrix)
//for the single-bin case
#pragma omp parallel for simd reduction(+:npairs)
for(int64_t i=0;I<nbuffer;i++) {
if(buffer[I] >= rmin && buffer[i] < rmax) npairs++;
}
And if we chose to go down the intrinsic route for the histogram update, then there are possible optimisations for small number of histogram bins - this
This has, of course, had me thinking of all the potential optimisations - another could be to change the order in which cell-pairs are computed to maximise the amount of date re-use. For example, if cellB1 has a lot of particles, then we might want to compute all the cell-pairs that involve cellB1. Currently, we compute in the order the cell pairs are generated -- (A0,B0), (A0, B1), (A0, B2), (A0, B-1), (A0, B-2), (A1, B0), (A1, B1) ... and so on. If we had a algorithm (really a cost function to optimise the ordering), then we could re-order the cells to improve cache re-use.
True, there might be an efficiency gain from larger histogramming chunks. If we want to optimize the histogram update for the case that ravg isn't needed, then the hybrid lookup table approach from the FCFC paper (section 3.2.3) may be worth testing. When ravg is needed, then the best optimization is probably to finish merging the linearbinning branch.
But if we're talking about all potential optimizations, then we should explore using cells smaller than Rmax! Right now, we count pairs in volume 18*Rmax^3 for each particle (18 instead of 27 because of the min_z_sep optimization). In the limit of infinitely small cells, we get to skip any that fall outside of the Rmax sphere, so the pair-counting volume becomes 4*pi*Rmax^3/3, or 4x smaller. Obviously the sweet spot for efficiency will not be the smallest cells possible, and will depend on the particle density, but the potential gain is substantial.
I will check out the hybrid LUT. Though my experience with the linear binning suggests that the bottleneck is not at the bin-index computing.
Not sure I follow the smaller cell size comment - isn’t that exactly what bin-refine-factor does? Or were you thinking of incorporating a tree-like spatial partitioning scheme?
I came across your numba_2pcf - looks very good 🙂! How hard would it be to implement the gridlink_in_ra_dec/gridlink_in_dec within that?
Ohhh and forgot about the one optimisation that SWIFT uses - projecting the points along the line joining the cell centers and removing all points that cannot be within Rmax. From what I recall, the paper said that that projection+cull works great for the shared corner/edge cell-pairs
I will check out the hybrid LUT. Though my experience with the linear binning suggests that the bottleneck is not at the bin-index computing.
That's what I would have thought too, but the benchmarks in that paper show that the histogram update is substantially faster in their code with a (hybrid) LUT. So it may be worth checking our code too.
Not sure I follow the smaller cell size comment - isn’t that exactly what bin-refine-factor does? Or were you thinking of incorporating a tree-like spatial partitioning scheme?
Maybe I misunderstood what bin_refine_factor was doing... I remember we talked about this on Zoom and I went away understanding that we weren't taking advantage of this! So we are actually discarding cell pairs if their minimum separation is greater than Rmax?
I came across your numba_2pcf - looks very good slightly_smiling_face! How hard would it be to implement the gridlink_in_ra_dec/gridlink_in_dec within that?
Thanks! Probably pretty easy; the code assumes 3D (x,y,z) inputs right now, but it shouldn't be too hard to relax that assumption.
Ohhh and forgot about the one optimisation that SWIFT uses - projecting the points along the line joining the cell centers and removing all points that cannot be within Rmax. From what I recall, the paper said that that projection+cull works great for the shared corner/edge cell-pairs
Yes, makes sense!
I will check out the hybrid LUT. Though my experience with the linear binning suggests that the bottleneck is not at the bin-index computing.
That's what I would have thought too, but the benchmarks in that paper show that the histogram update is substantially faster in their code with a (hybrid) LUT. So it may be worth checking our code too.
Huh - do you know what they did?
Not sure I follow the smaller cell size comment - isn’t that exactly what bin-refine-factor does? Or were you thinking of incorporating a tree-like spatial partitioning scheme?
Maybe I misunderstood what bin_refine_factor was doing... I remember we talked about this on Zoom and I went away understanding that we weren't taking advantage of this! So we are actually discarding cell pairs if their minimum separation is greater than Rmax?
Yup. Though that does not happen often for small bin-ref. Allowing bin-ref to increase arbitrarily would then hit the hard NLATMAX limit we have coded in.
We could use yield individual cell-pairs from within the generate-cell-pairs function and then use OMP tasks to process each cell-pair. That would remove the memory footprint of the cell-pairs and we could really take advantage of the smaller cells.
I came across your numba_2pcf - looks very good slightly_smiling_face! How hard would it be to implement the gridlink_in_ra_dec/gridlink_in_dec within that?
Thanks! Probably pretty easy; the code assumes 3D (x,y,z) inputs right now, but it shouldn't be too hard to relax that assumption.
If you are feeling up to it and implement those gridding functions in numba-2pcf, let know. Think the dot product way of computing angular correlation functions will be much faster than Corrfunc.
Do you also have a handy utility function in there to generate cell-pairs from a set of two cells and a neighborhood window?
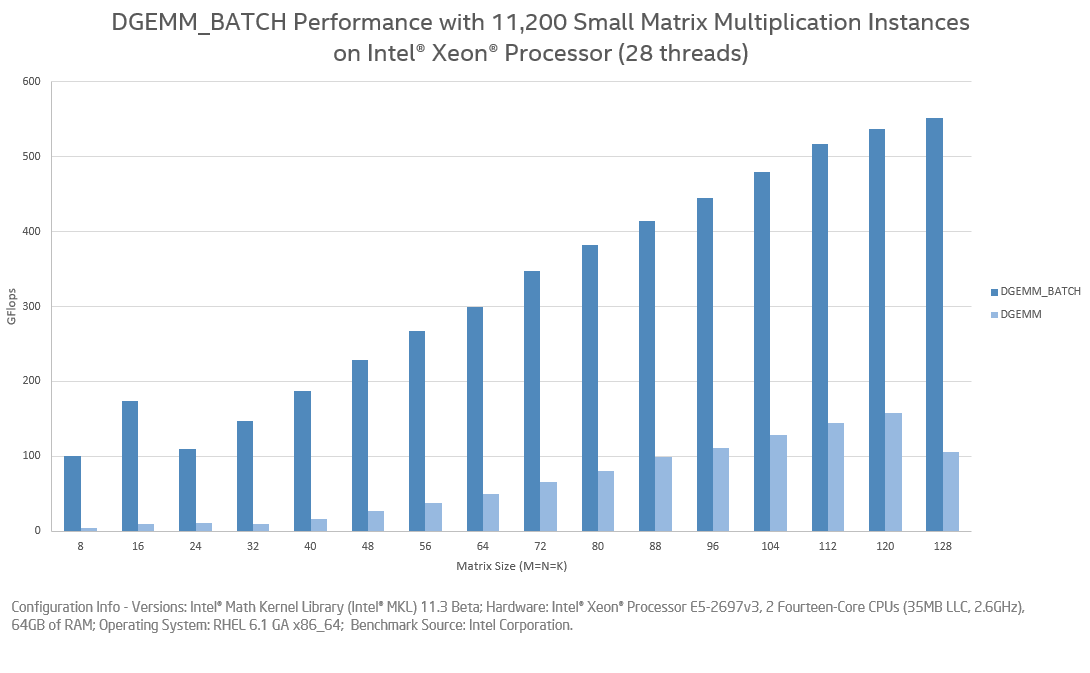
In case I forget, MKL has a batch ?GEMM feature that can "N" multiplications of small matrices in a single call and is much more efficient than "N" calls to individual ?GEMMs. More details here
Huh - do you know what they did?
I think it's all in their paper; I haven't investigated beyond that. Looks like the source code is here.
We could use yield individual cell-pairs from within the generate-cell-pairs function and then use OMP tasks to process each cell-pair. That would remove the memory footprint of the cell-pairs and we could really take advantage of the smaller cells.
Agreed, OpenMP tasks could be good for this!
If you are feeling up to it and implement those gridding functions in numba-2pcf, let know. Think the dot product way of computing angular correlation functions will be much faster than Corrfunc.
I probably won't have time to do this myself, but I'm happy to answer questions.
Do you also have a handy utility function in there to generate cell-pairs from a set of two cells and a neighborhood window?
The code just loops over the neighborhood window rather than pre-generating the list of cell pairs, but switching to a pre-generation method wouldn't be hard.
Just an update from the experiments - the overall runtime is not competitive with Corrfunc. In my experiments, I did the entire the cell-pair (i.e., computed all distances between all possible particle pairs) distances and stored into a malloc'ed buffer, and then did the histogram update. However, the DGEMM runtime was not competitive with Corrfunc for cases with small $\theta{max}$. In cases where $\theta{max} >= 60$ degrees, then the DGEMM runtime is better than Corrfunc. I only tried with the Apple Accelerate BLAS and not OpenBlas or MKL. I will put my experiments up on a gist and link that to here later
For cases where neither
rpavgandweightavghave been requested, using a DGEMM matrix-multiplication would speed up the code significantly. This is particularly true forDDtheta_mockswhere a DGEMM enabled $X1 \cdot X2$ can be used to compute $cos(\theta)$ directly.This requires adding another
Npartloop withingridlink_*to compute the actual cell occupancy and then malloc'ing the X array to be 3 times the size, and defining the Y and Z arrays to be offset from the X array (i.e,lattice[I].y = lattice[I].x + lattice[i].nelements && lattice[I].z = lattice[i].x + 2*lattice[i].nelements. This strategy will only work withcopy_particles=TrueA nice side-effect will be that all of the code around
realloccan be removed. There will certainly be a performance hit but hopefully not too major (the loop can be OpenMP parallelised trivially)@lgarrison What do you think?