Yes, happy to help and/or for my code to be used. A couple early thoughts:
- I've made the code much more robust and put it in a R package (thought it would spruce up my RStudio table contest submission😃), repo here: https://github.com/rdboyes/forestable
- Re: screwy viewer alignment - my current solution is to save the image, then display it to the Viewer pane using a call to plot(magick::image_read(file_path)). It's a little hacky but it works for now. Inside RMarkdown, the tables can be displayed with a call to knitr::include_graphics, which seems to work locally but not in github readmes for some reason.


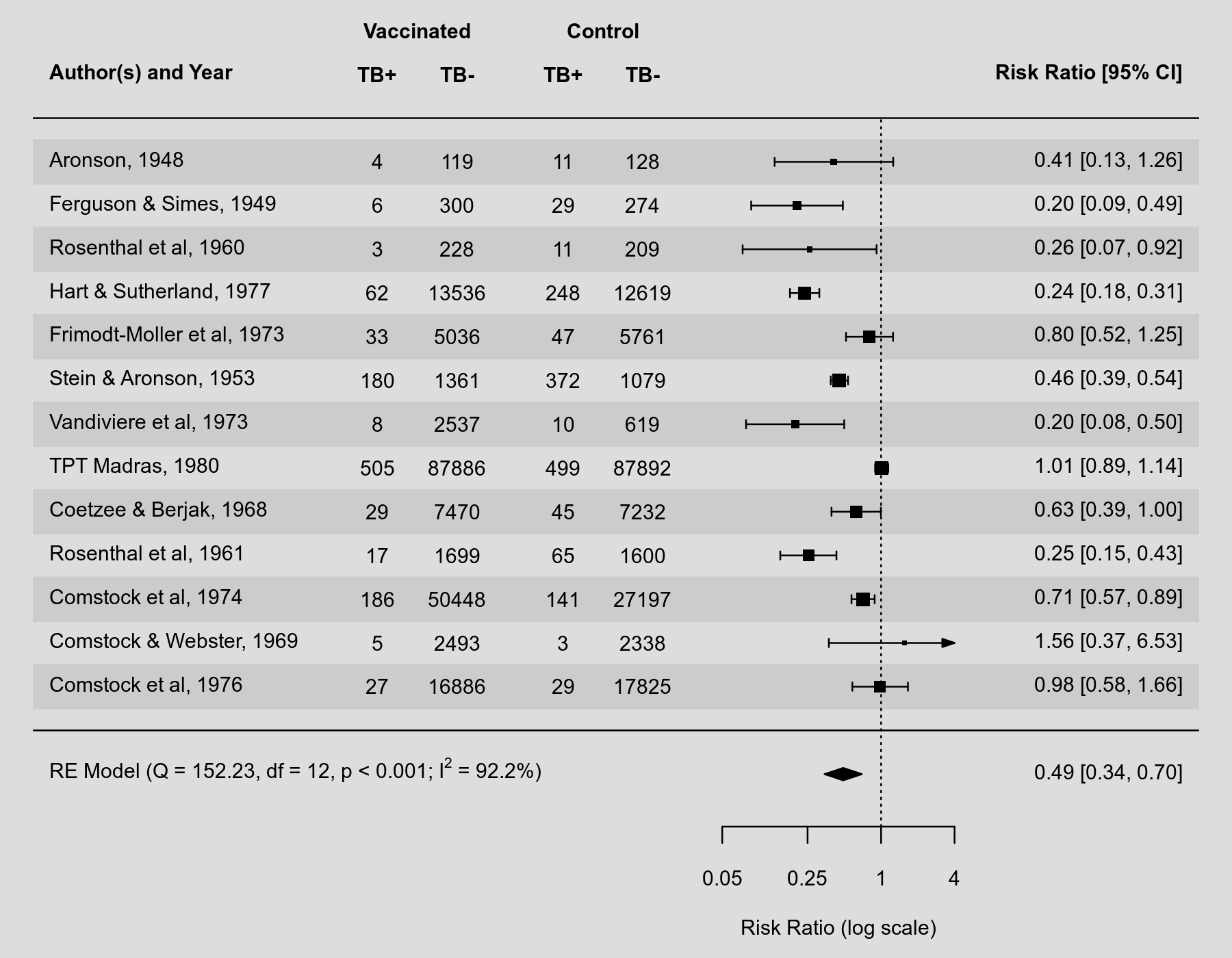











Problem The most recent paper on ROB2, and indeed the Cochrane Handbook itself, suggest presenting risk-of-bias information as part of a paired forest plot. Risk-of-bias assessments are intended to be result-specific, and so presenting information about the potential bias in a result alongside the main analysis helps to ensure that the results are interpreted appropriately. The paired figure from the ROB2 paper is below, for reference:
This plot is something that has been requested by other users and has been on the cards for a while:
Until recently, I was not sure how to do it, as the
metafor::forest()function is quite restrictive in terms of matching with anything else, and aligning text tables inggplot2is a pain. However, @rdboyes recently shared code for producing a beautiful forest plot to Twitter. The code is available here: https://github.com/rdboyes/Forest_plot_with_table/blob/master/create_figure.RDescribe the solution you'd like Ideally, I'd like to add a function (
rob_blobbogram()) torobviswhich creates these table/forest-plots/risk-of-bias plots.The function would take two main inputs:
rmaobject frommetafor::rma())rob_*()functions (for consistency)Plus then some styling arguments, similar to the other
rob_*()functions.Describe alternatives you've considered I have looked at packages like
patchworkandcowplotto see if two disctinct plots could be aligned easily. It is possible - howeverggplot2is not great at handling text/tables. So while the forest plot and risk-of-bias plots could be aligned, adding the extra tables describing the study's results proved difficult.Predicted issues One important issue is that when previewing the plot created by @rdboyes, it looks scewy in the RStudio viewer but perfect once saved to a file. From a user interface point of view, this will cause a lot of issues as people email to say the graph looks wrong. I think it should be possible to address this by setting the dimensions of the RStudio viewer panel, but I haven't looked into it a whole lot. Perhaps @rdboyes, you might know?
Secondly, the current code for the forest-plot draws from a table rather than a meta-analysis object, and also does not include other aspects produced by the meta-analysis such as overall summaries or heterogeneity. However, by adding rows containing these, it should be possible to adapt the existing code to work.
Practical issues
@AJFOWLER - are you interested in having a go at this, as it might be a bit more interesting/challenging for you than refactoring the current code base?
@rdboyes - are you happy for us to reuse your forest plot code here/be involved in developing this functionality?