As explained in the tutorial, you can make selections through the interface to select gene clusters of interest:
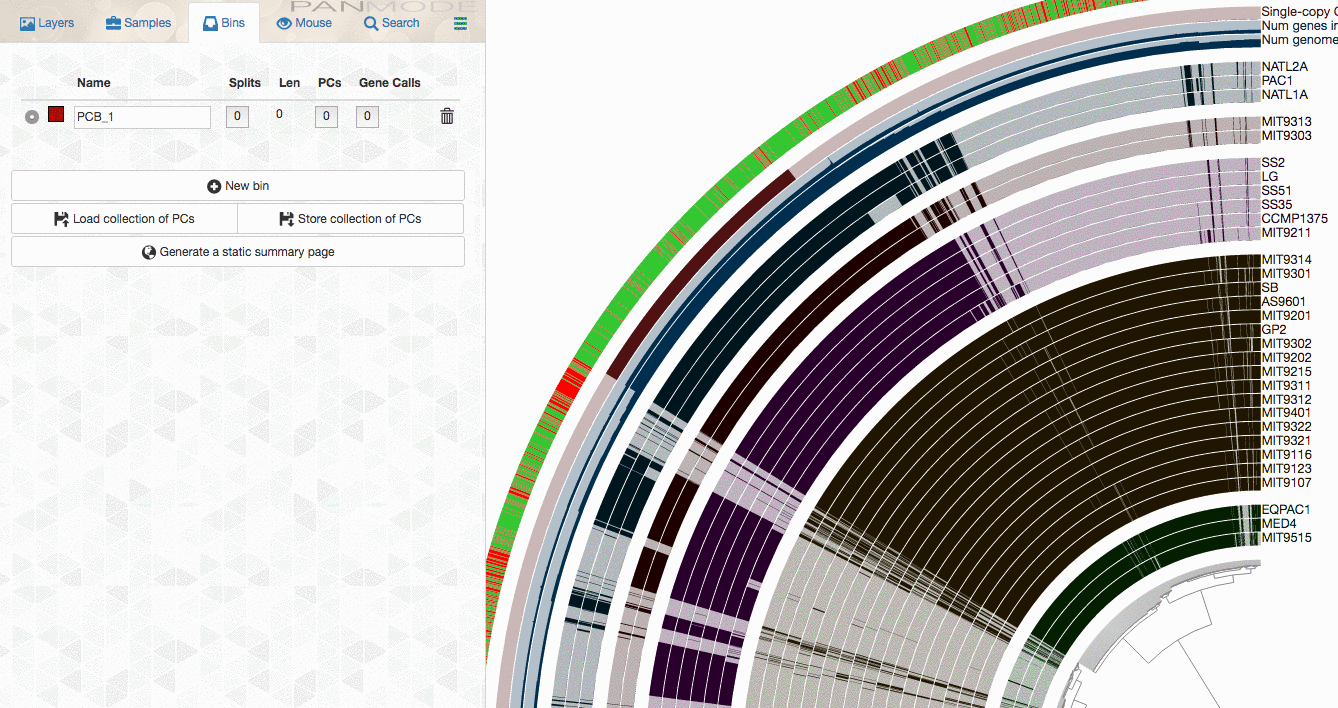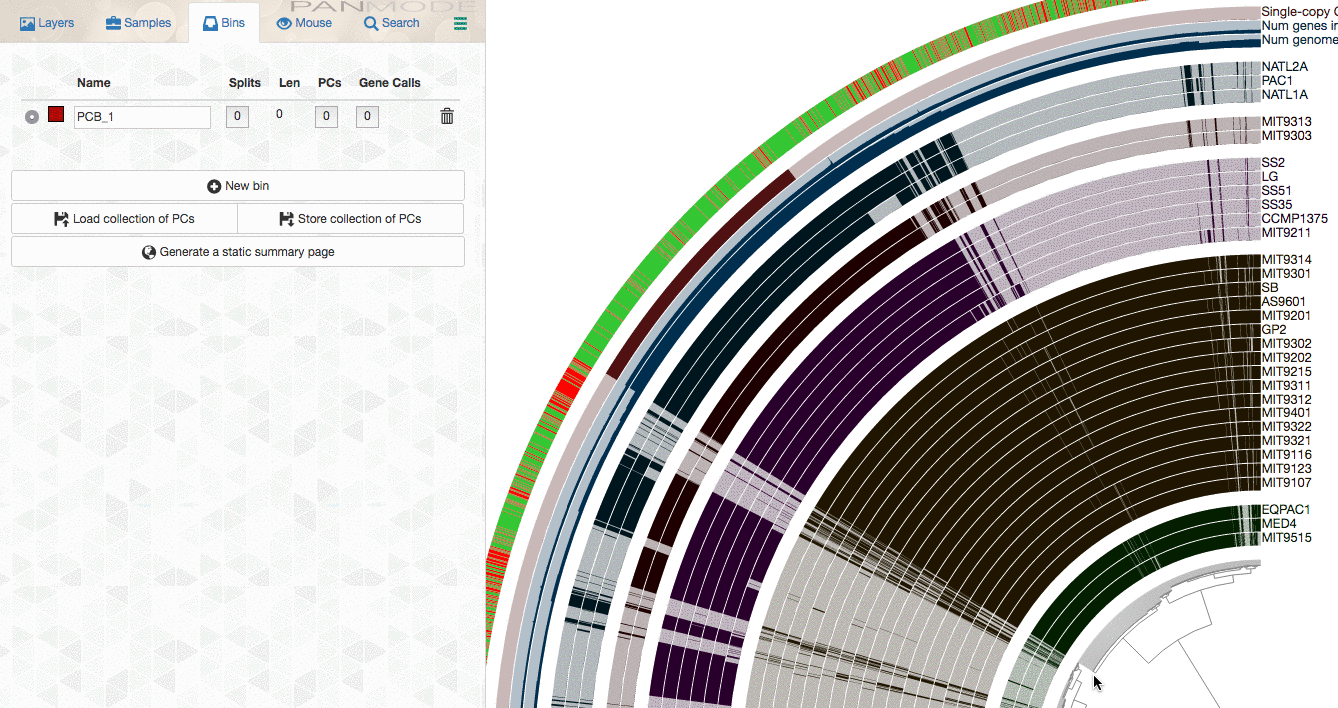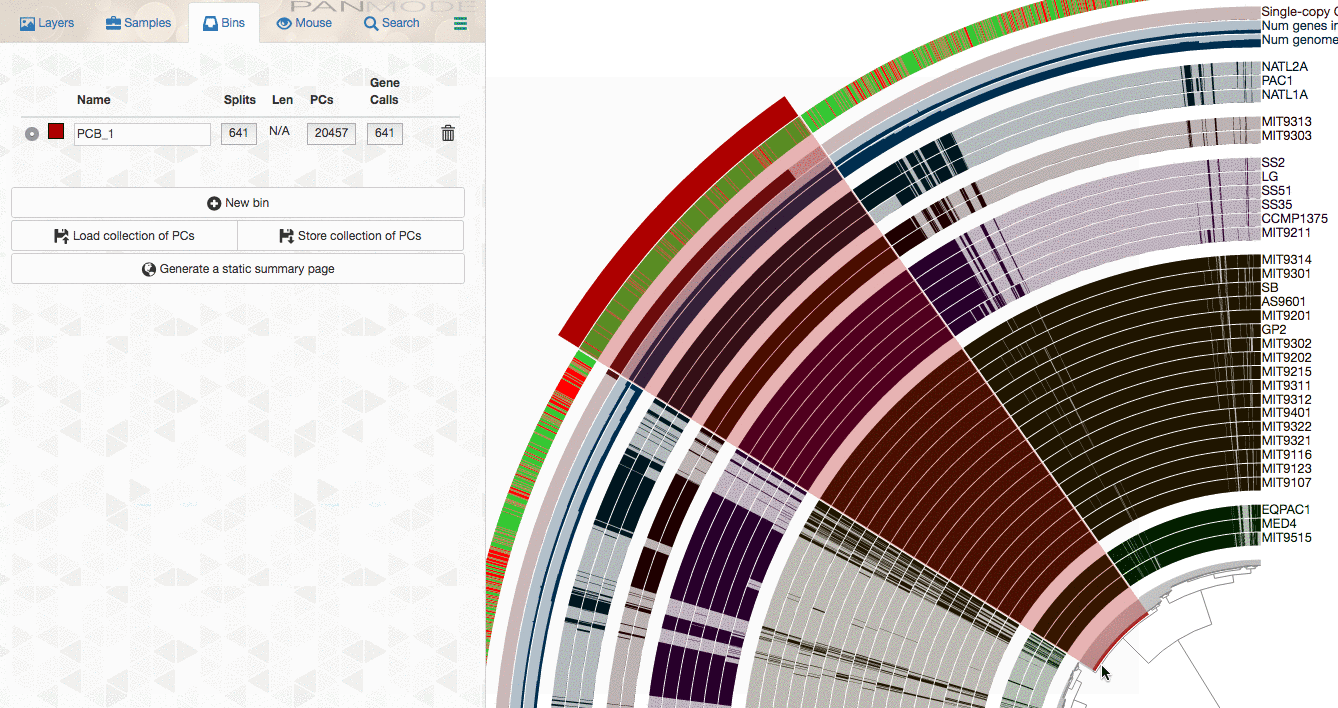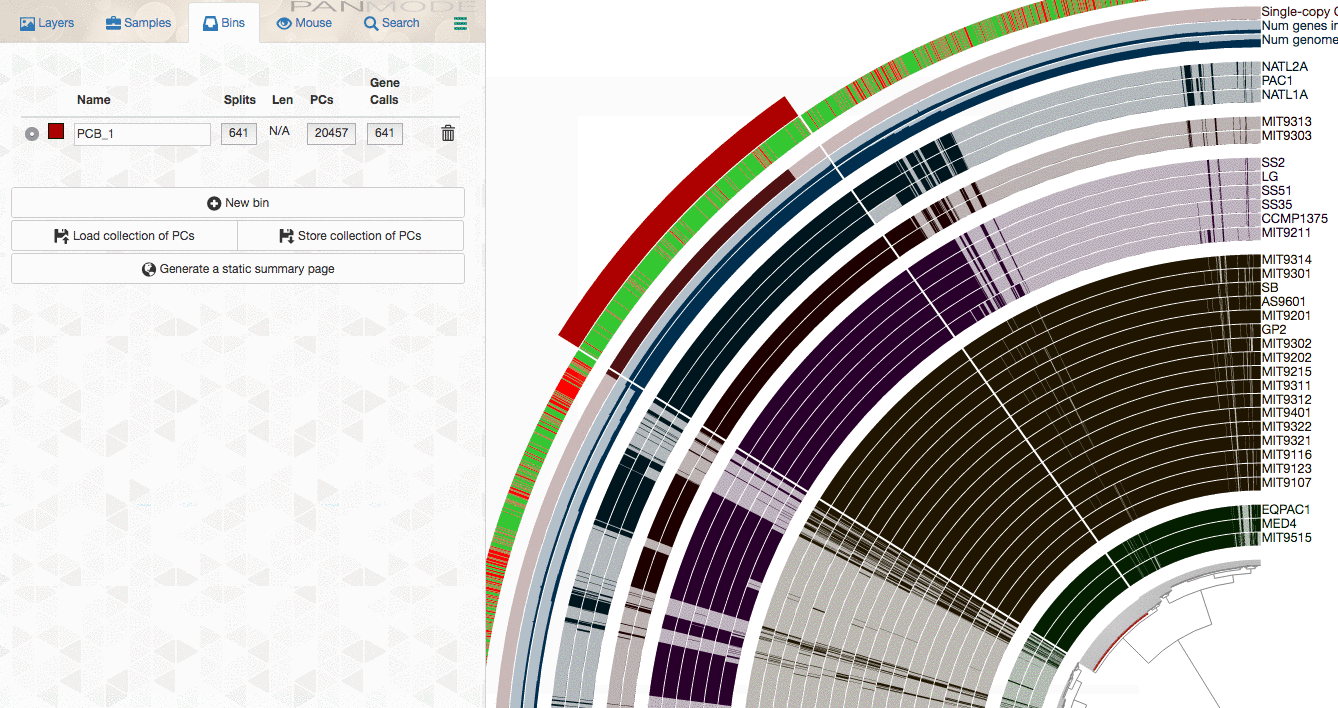
or use more advanced filters to identify gene clusters to focus, then store them as a collection, and use anvi-summarize to get the summary output to access to all gene clusters of interest along with their attributes.
I have a question about the pangenomics workflow:
If I look at the image from the phylogenome workflow (see below), I see the regions that show high similarity.
How can I extract these regions from all of those bacteria (I want to compare the gene clusters in this regions).
Thank you for your help.