Inferring taxonomy for metagenomes
[!NOTE] The names in the output of the following two commands is not the same as on the tutorial page
Ribosomal_S6_2915(Tutorial)Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_11655(my output).
command
anvi-estimate-scg-taxonomy -c CONTIGS.db \
--metagenome-mode```output
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| | percent_identity | taxonomy |
+==========================================================+====================+=================================================================================================================================+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_11655 | 98.9 | Bacteria / Firmicutes / Bacilli / Staphylococcales / Staphylococcaceae / Staphylococcus / Staphylococcus epidermidis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_12163 | 98.9 | Bacteria / Firmicutes / Bacilli / Staphylococcales / Staphylococcaceae / Staphylococcus / Staphylococcus hominis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_15200 | 98.9 | Bacteria / Firmicutes / Bacilli / Lactobacillales / Lactobacillaceae / Leuconostoc / Leuconostoc citreum |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_25880 | 98.9 | Bacteria / Firmicutes / Clostridia / Tissierellales / Peptoniphilaceae / Finegoldia / |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_2915 | 97.9 | Bacteria / Firmicutes / Bacilli / Lactobacillales / Enterococcaceae / Enterococcus / Enterococcus faecalis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_29818 | 97.9 | Bacteria / Firmicutes / Bacilli / Lactobacillales / Streptococcaceae / Streptococcus / |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_30904 | 98.1 | Bacteria / Firmicutes / Clostridia / Tissierellales / Peptoniphilaceae / Anaerococcus / Anaerococcus nagyae |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_4484 | 98.9 | Bacteria / Firmicutes / Clostridia / Tissierellales / Peptoniphilaceae / Peptoniphilus / Peptoniphilus lacydonensis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_6421 | 100 | Bacteria / Actinobacteriota / Actinomycetia / Propionibacteriales / Propionibacteriaceae / Cutibacterium / Cutibacterium avidum |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_7660 | 98.9 | Bacteria / Firmicutes / Bacilli / Staphylococcales / Staphylococcaceae / Staphylococcus / |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+command
anvi-estimate-scg-taxonomy -c CONTIGS.db \
> -p PROFILE.db \
> --metagenome-mode \
> --compute-scg-coveragesoutput
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| | percent_identity | taxonomy | DAY_15A | DAY_15B | DAY_16 | DAY_17A | DAY_17B | ... 6 more |
+==========================================================+====================+================================+===========+===========+==========+===========+===========+==============+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_2915 | 97.9 | (s) Enterococcus faecalis | 372.512 | 699.853 | 663.241 | 186.34 | 1149.6 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_11655 | 98.9 | (s) Staphylococcus epidermidis | 112.694 | 172.478 | 147.304 | 23.3901 | 140.769 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_4484 | 98.9 | (s) Peptoniphilus lacydonensis | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_7660 | 98.9 | (g) Staphylococcus | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_6421 | 100 | (s) Cutibacterium avidum | 17.8935 | 5.87368 | 4.79909 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_12163 | 98.9 | (s) Staphylococcus hominis | 2.39322 | 22.5447 | 13.2806 | 0 | 9.94853 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_15200 | 98.9 | (s) Leuconostoc citreum | 0 | 0 | 1.86532 | 0 | 1.6936 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_29818 | 97.9 | (g) Streptococcus | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_25880 | 98.9 | (g) Finegoldia | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_30904 | 98.1 | (s) Anaerococcus nagyae | 0.528958 | 0 | 0.14157 | 0 | 0 | ... 6 more |
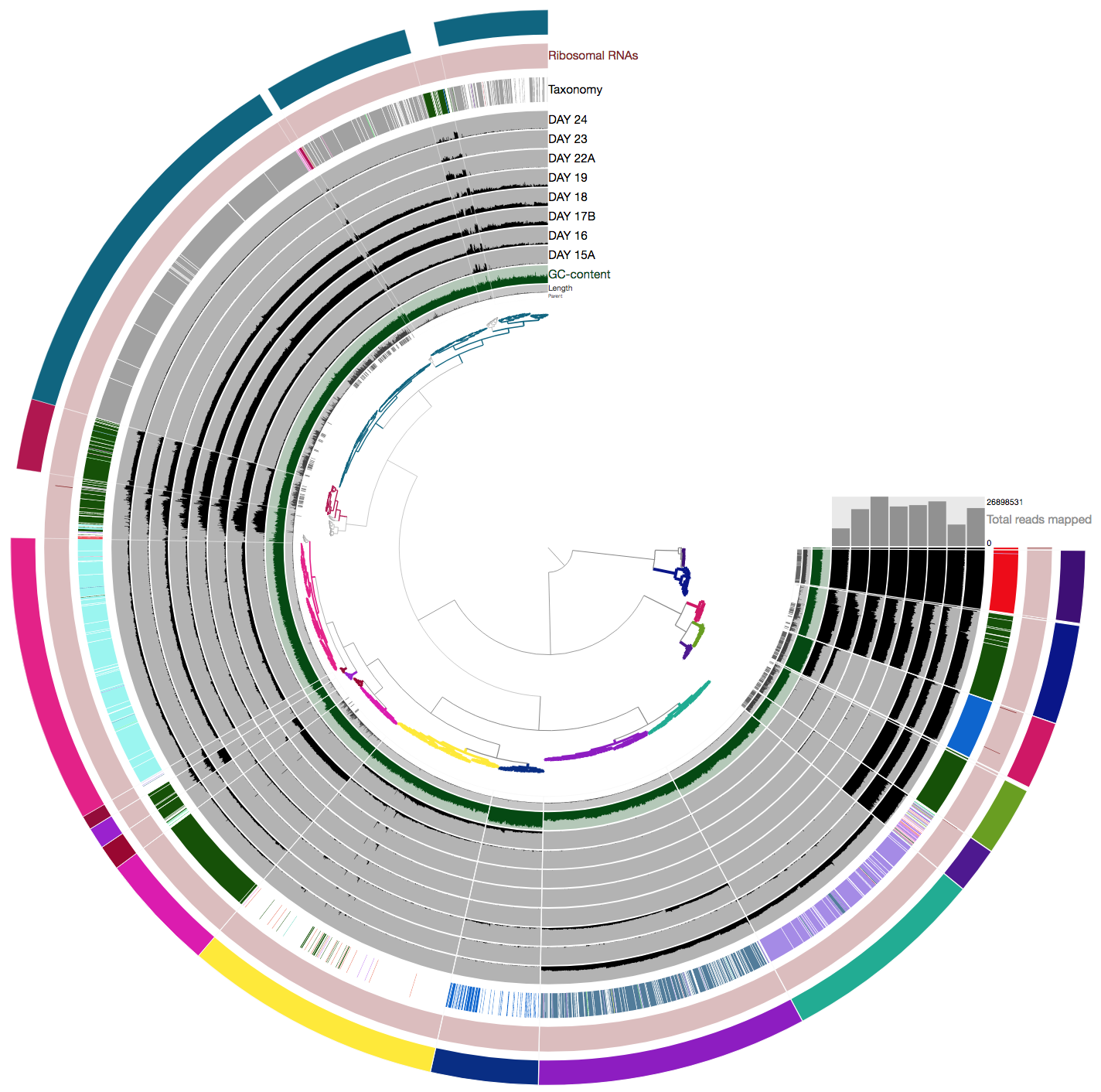
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
Collection of discrepancies I noticed between the tutorial (after Mete's updates) and my outputs. I'll write one comment per issue as I notice them.