test file:
### Fig.1
- ### Overview of SIFT.
- 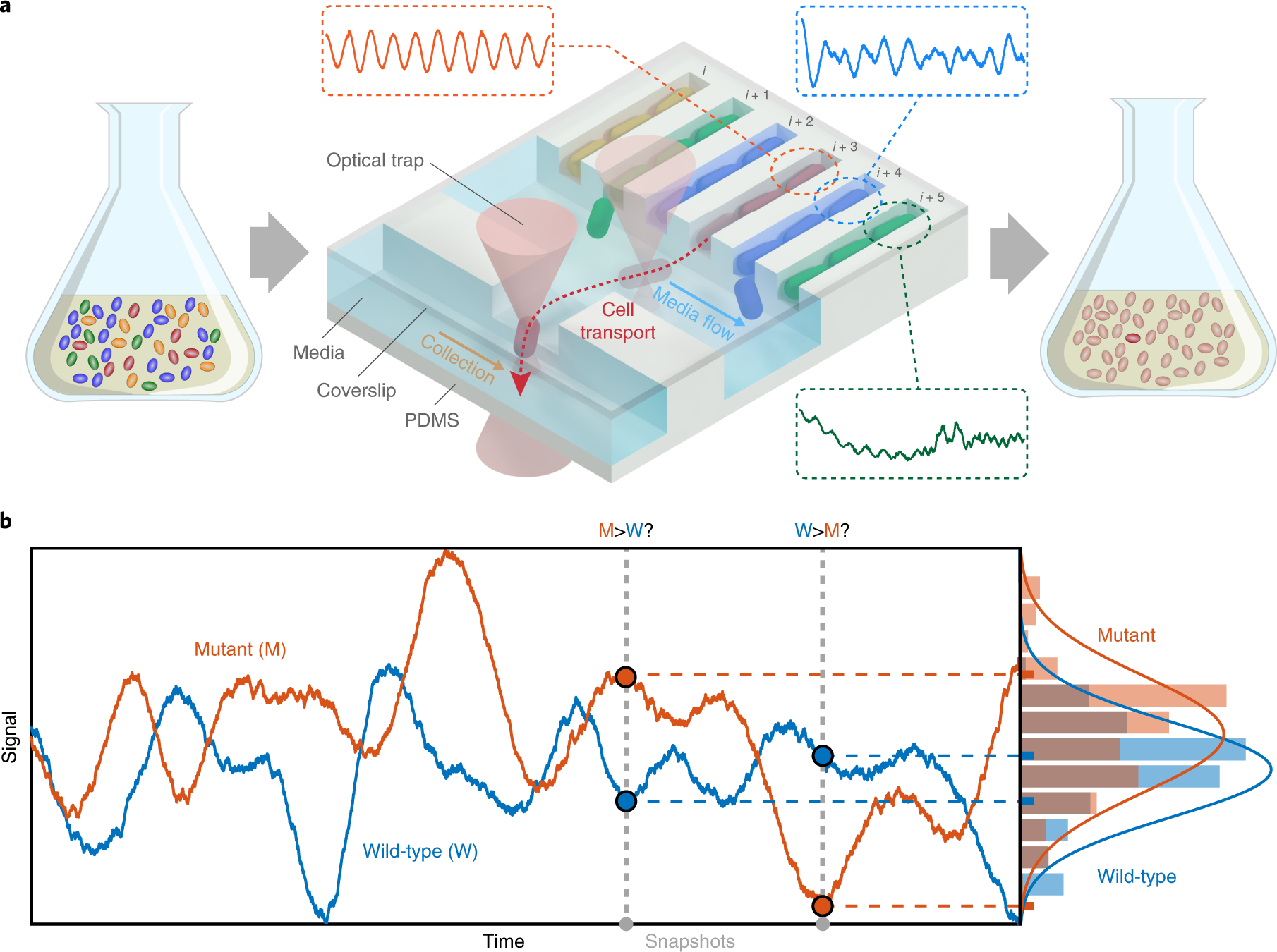
- `a` | Schematic of the screening process.
- Any *pooled library or complex culture* (mixture of colored ovals)
- can be
- loaded into the microfluidic device and
- characterized by long-term time-lapse microscopy.
- After *phenotyping*,
- cells of interest (red cylinders) are
- transported by optical trapping to a clean area on the chip
- -- previously sterilized with the support of a series of push-down valves --
- and individually collected from the device without cross-contamination.
- Isolated cells may be propagated for **any** downstream off-chip analysis.
- `b`
- A hypothetical example of two different genotypes
- (represented by generic wild-type (`W`) and mutant (`M`) forms)
- that are frequently mischaracterized by individual single-cell snapshots (colored circles).
- Only
- by estimating the *full distributions of the phenotypic property* (right panel),
- by assaying the same genotypes over *long periods of time* (left panel),
- are the correct classifications reliably assigned.
### Fig.2
- ### Cell retrieval principles of the SIFT platform.
- 
- `a`
- A macroscopic image of the entire microfluidic chip,
- loaded with dyes for visualization,
- is shown in the `left panel`.
- `Dotted boxes` correspond to regions represented in other panels.
- `Scale bar`, 5 mm.
- A schematic overview of the single-cell isolation process
- is shown in the `right panel`,
- corresponding
- to a not-to-scale representation
- of the `blue boxed region` from the left panel.
- Cell(s) from a lineage of interest
- (marked by `red star`)
- are optically trapped and transported to a *clean section of the chip*
- -- segregated by a series of **collection valves** (`green box`) --
- for further *characterization* and/or immediate *collection*.
- `b` | Kymograph of cell transportation via *optical trapping*.
- An $Escherichia~ coli$ cell of interest
- is
- removed from a growth trench (leftmost two panels) and
- *dragged through* an open **collection valve** (rightmost two panels) for off-chip collection.
- - `White circles` mark the optical trap position;
- `dotted arrows` indicate the movement direction of the trapped cell.
- Similar cell transfers were *performed more than 200 times across 5 independent screening runs* with similar results. Scale bar, 5 µm.
- `c` | On-chip inlet-cleaning valve infrastructure.
- A top-down representation of
- the entrances for one *microfluidic lane*
- (`left panel`; magnified representation of the orange dotted box from the left panel of a)
- shows deposited bacterial biofilms
- cleaned by circulated bleach (`arrows`),
- while downstream cells residing in growth cavities
- are protected by a closed push-down valve (`red indented box`).
- A cross-sectional view of the *push-down valves*
- (`right panel`; magnified depiction of dotted grey box in the left panel) shows valve actuation.
- *Pressurization of a ceiling chamber* causes deflection of a thin PDMS layer that pinches flow channels (yellow) closed.
- `d`
- Images of a feeding lane inlet
- before (`left panel`) and after (`right panel`) biofilm removal
- by the chip-cleaning procedure outlined in `c`.
- Similar results were observed across five independent screening runs. Scale bar, 25 µm.
### Fig.3
- ### Clean, reliable isolation of live individual cells by SIFT.
- 
- `a`
- Sample fields-of-view from the mock color screen
- comprised of *overlaid phase contrast* and *fluorescence images*.
- $E.~ coli$ were
- engineered to constitutively express either red, yellow, cyan or no fluorescent protein (*RFP*, *YFP*, *CFP* or no *FP*), and
- mixed to a ratio of 1:1:1:100, respectively.
- Scale bar, 5 µm.
- `b`
- Collection of the chip flow-through
- prior to SIFT screening the mock library of mixed colored cells (*left panel*).
- Three out of three individual cells were isolated from each targeted lineage in the intended sequence.
- Each agar well is a plating of an independent collection bin; numbers denote the order of collection. Similar results were observed across three independent mock screening runs. Scale bars, 10 mm.
- `c`
- Number of genomic mutations specific to 15 isolated single cells transported via an optical trap for 15 s, 30 s and 75 s (the standard trapping time, two-times longer and five-times longer, respectively). All cells were continuously moved for the entire duration of the respective trapping times to mimic screening transportations, as shown in the schematic. Data were gathered from a single screening run.
- `d`
- Growth-rate-derived generation times immediately following optical transport for varying times, ranging from just over the standard trapping time (15 s) to greater than 5 times longer. Mean generation times (red circles) were measured over a 12-h observation period, with error bars as s.e.m. Mean generation time of 50 non-trapped cells (dotted line; μ) and twice the standard deviation (shaded region; 2σ) represent natural growth heterogeneity within the same experiment. Data were gathered from a single screening run.
### Fig.4
- ### Genetic screen of a dual-feedback oscillator library.
- 
- `a`
- The **dual-feedback oscillator** with linked positive and negative feedback loops (`top left panel`).
- All circuit genes,
- including the GFP reporter,
- were encoded
- on *two plasmids*, transcriptionally regulated by **identical $P_{lac/ara-1}$ hybrid promoters**, and
- marked for protein degradation by SsrA (LAA) tags in $E.~ coli$ (top right panel).
- Library mutations
- were targeted to *promoter, operator and RBS sites*
- of both $lacI$ and $araC^{const}$, the araC gene with a point mutation (Y13H) conferring constitutive activator functionality (bottom panel).
- `b`
- Statistical estimates of period CV and mean period of all library variants with oscillatory-like signals over a 24-h period using SIFT (Nosc = 7,803).
- Variants selected to highlight phenotypic diversity (`grey circles` with `roman numerals`) have time-traces shown in `c`.
- `Red circles` represent variants that were isolated, some of which were individually characterized in a follow-up mother machine run.
- Mean generation time was 25 min. Library phenotype data were gathered from a single screening run.
- `c`
- Time-traces of selected variants, as described in `b`.
- `Orange dots` indicate called peaks.
- `<T>`, mean period; `CVT`, period CV; `a.u.`, arbitrary units.
- `d`
- Representative time-traces of the original dual-feedback circuit (SL126) and the best-performing (that is, with the lowest period CV) isolated mutant (SL278), characterized in the mother machine under conditions without any supplemented IPTG or arabinose.
more:
Steps to Reproduce:
"editor.cursorSurroundingLines": 50.mdfile with several picturesUPandDOWNto read or edit the file, the page will jump randomly...previewpage with two fingers. When a second picture raise from bottom, the first picture suddenly jumped back.Does this issue occur when all extensions are disabled?: Yes/No