Current default threshold is 0.25 for mne.preprocessing.ICA.find_bads_ecg with method='ctps'.
However, the value seems arbitrary. Would it make sense to implement automatic computation for CTPS threshold?
0.25 is actually the threshold for data with 400 Hz sampling rate. I found that 0.25 is sometimes not sensitive enough for clean data with sampling rate higher than 400 Hz.
For example, the sampling rate of sample dataset is 600 Hz. After filtering, mne.preprocessing.ICA.find_bads_ecg does not detect any ECG related IC using default (0.25) threshold. Using 0.21 (computed from the formula), it is able to detect one ECG related IC.
The ECG Evoked
The ECG scores of ICs
The detected IC (ICA001) with threshold=0.21
Describe your solution
In the reference, the threshold was set to Pk >= 10^-20. Pk can be computed from formula (13) with λ and λ will change according to the number of data points. Thus, the threshold will also change according to the sampling rate.
The threshold of the Kuiper index can be determined easily:
import numpy as np
def _get_ctps_threshold(N=1000, Pk_threshold=1e-20):
vs = [x / 100 for x in range(1, 100)]
Pks = list()
C = np.sqrt(N) + 0.155 + 0.24 / np.sqrt(N)
# because when k gets large, only k=1 matters for the summation
# k*v*C thus becomes v*C
for v in vs:
Pk = 2 * (4 * (v * C)**2 - 1) * (np.exp(-2 * (v * C)**2))
Pks.append(abs(Pk - Pk_threshold))
return vs[Pks.index(min(Pks))]
In [2]: _get_ctps_threshold(400)
Out[2]: 0.25
In [3]: _get_ctps_threshold(600)
Out[3]: 0.21
In [4]: _get_ctps_threshold(1000)
Out[4]: 0.16
If that's reasonable, I can make a PR for this implementation.
Additional context
The test script:
import os.path as op
from IPython import get_ipython
import mne
from autoreject import get_rejection_threshold
mne.set_log_level('INFO')
get_ipython().run_line_magic('matplotlib', 'qt')
get_ipython().run_line_magic('gui', 'qt')
h_freq = None
l_freq = 40
# Calibration files for Maxwell filter
sample_dir = mne.datasets.sample.data_path()
ctc = op.join(sample_dir, 'SSS', 'ct_sparse_mgh.fif')
cal = op.join(sample_dir, 'SSS', 'sss_cal_mgh.dat')
raw_fname = op.join(sample_dir, 'MEG', 'sample', 'sample_audvis_raw.fif')
sss_fname = op.join(sample_dir, 'MEG', 'sample', 'sample_audvis_raw_sss.fif')
raw = mne.io.read_raw_fif(raw_fname, preload=True)
# remove bad channels
raw.info['bads'].extend(['MEG 1032', 'MEG 2313'])
raw_sss = mne.preprocessing.maxwell_filter(raw,
calibration=cal,
cross_talk=ctc)
# filter
raw_sss.filter(None, 40)
# ICA
n_components = 0.999
ica = mne.preprocessing.ICA(n_components=n_components, random_state=99)
picks_eog = mne.pick_types(raw.info,
meg=True,
eeg=False,
eog=False,
stim=False,
exclude='bads')
# high-pass for ICA
raw_sss.filter(1, None, picks=picks_eog, fir_window='hann')
# Only remove ECG artifacts for now
picks = mne.pick_types(raw.info,
meg=True,
eeg=False,
eog=False,
stim=False,
exclude='bads')
# use autoreject to find the rejection threshold to get better ICA results
tstep = 1.0
events = mne.make_fixed_length_events(raw_sss, duration=tstep)
even_epochs = mne.Epochs(raw_sss,
events,
baseline=(0, 0),
tmin=0.0,
tmax=tstep,
reject_by_annotation=True)
reject = get_rejection_threshold(even_epochs,
ch_types=['mag', 'grad'])
ica.fit(raw_sss, picks=picks, reject=reject, tstep=tstep)
# ECG
ecg_epochs = mne.preprocessing.create_ecg_epochs(raw_sss)
ecg_epochs.apply_baseline((None, None))
# threshold = 0.25
# found 0 components
ecg_inds, ecg_scores = ica.find_bads_ecg(ecg_epochs,
method='ctps',
threshold=0.25)
ecg_evoked = ecg_epochs.average()
ecg_evoked.plot()
# barplot of ICA component "ECG match" scores
ica.plot_scores(ecg_scores)
print(ecg_inds)
# threshold = automatically for 600Hz data
# Using threshold: 0.21 for CTPS ECG detection
# found 1 components [1]
ecg_inds, ecg_scores = ica.find_bads_ecg(ecg_epochs, method='ctps')
print(ecg_inds)
# plot diagnostics
ica.plot_properties(raw_sss, picks=ecg_inds)
# plot ICs applied to raw data, with ECG matches highlighted
ica.plot_sources(raw_sss)
# plot ICs applied to the averaged ECG epochs, with ECG matches highlighted
ica.plot_sources(ecg_evoked)
ica.plot_overlay(raw_sss, exclude=ecg_inds, picks='mag')
Describe the problem
Current default threshold is 0.25 for
mne.preprocessing.ICA.find_bads_ecgwithmethod='ctps'. However, the value seems arbitrary. Would it make sense to implement automatic computation for CTPS threshold?0.25 is actually the threshold for data with 400 Hz sampling rate. I found that 0.25 is sometimes not sensitive enough for clean data with sampling rate higher than 400 Hz.
For example, the sampling rate of
sampledataset is 600 Hz. After filtering,mne.preprocessing.ICA.find_bads_ecgdoes not detect any ECG related IC using default (0.25) threshold. Using 0.21 (computed from the formula), it is able to detect one ECG related IC.The ECG Evoked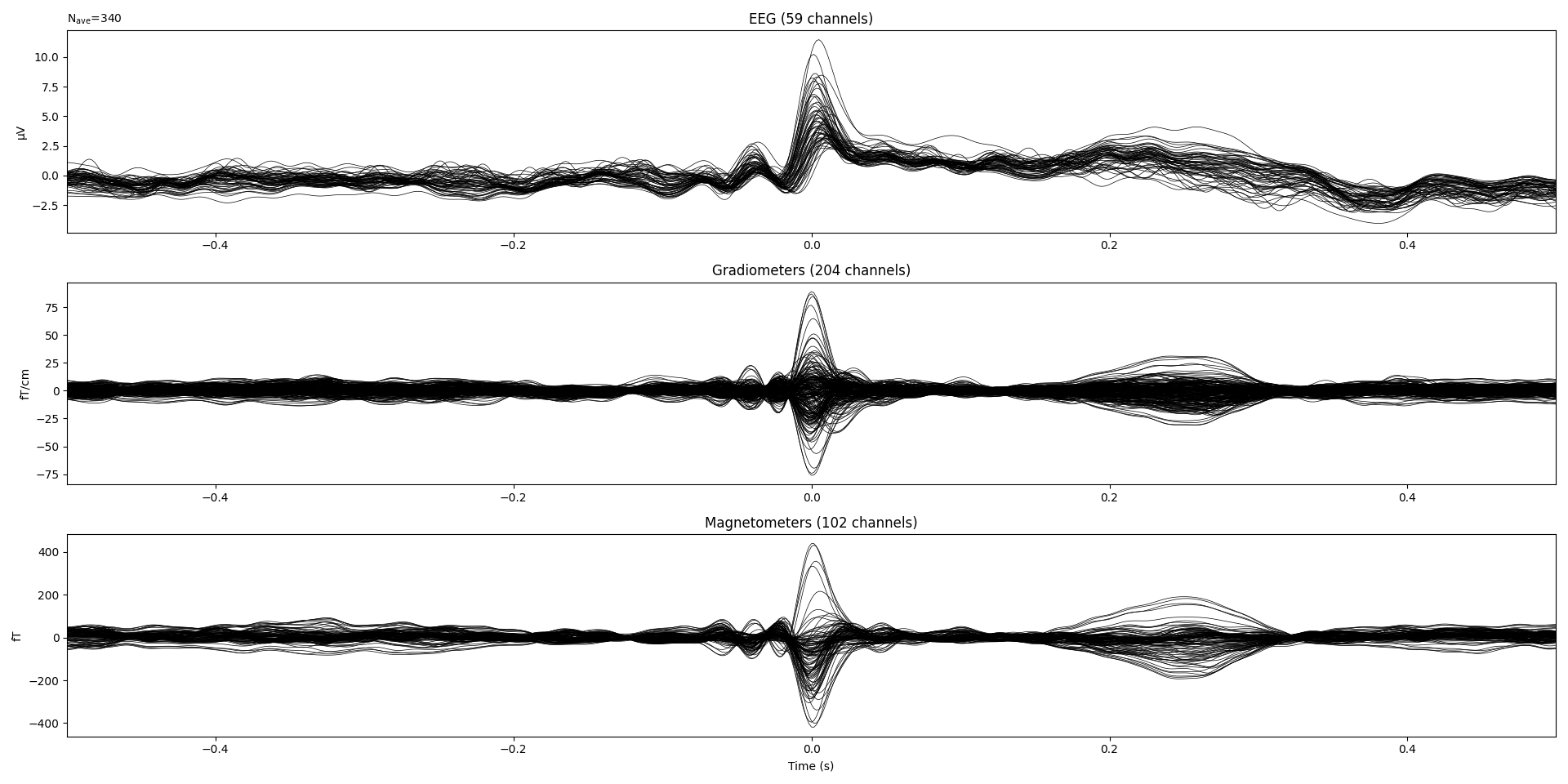
The ECG scores of ICs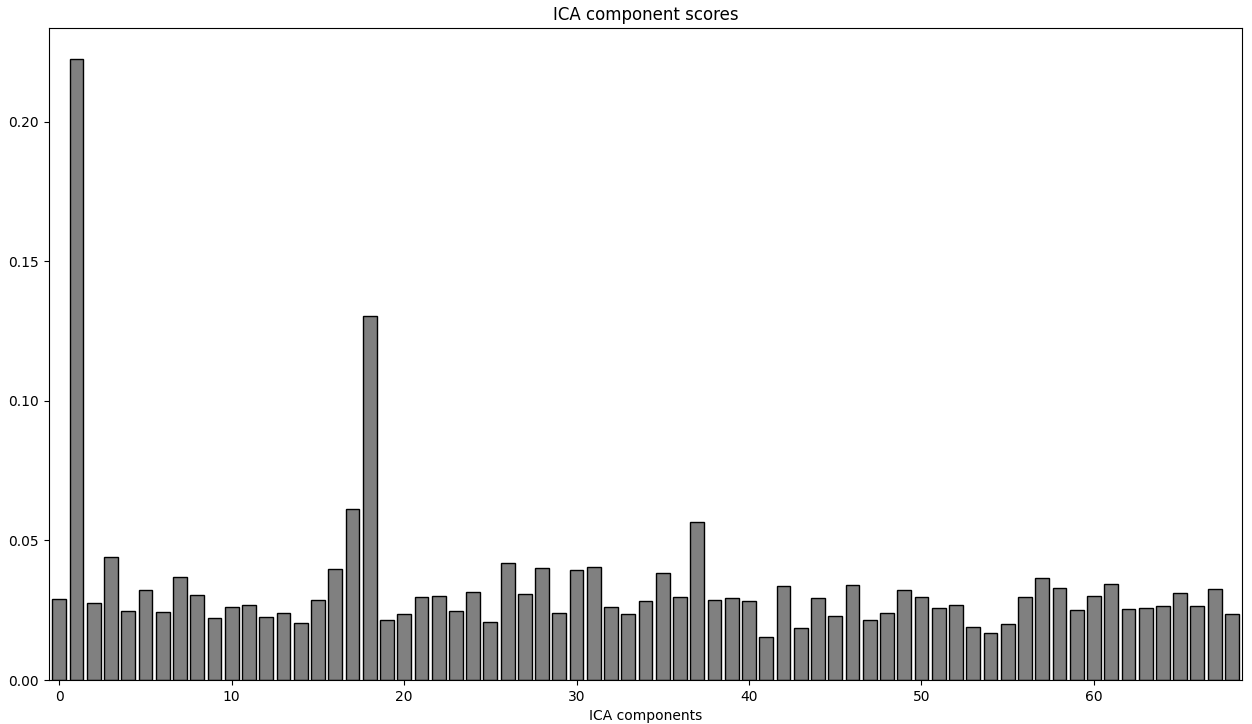
The detected IC (ICA001) with

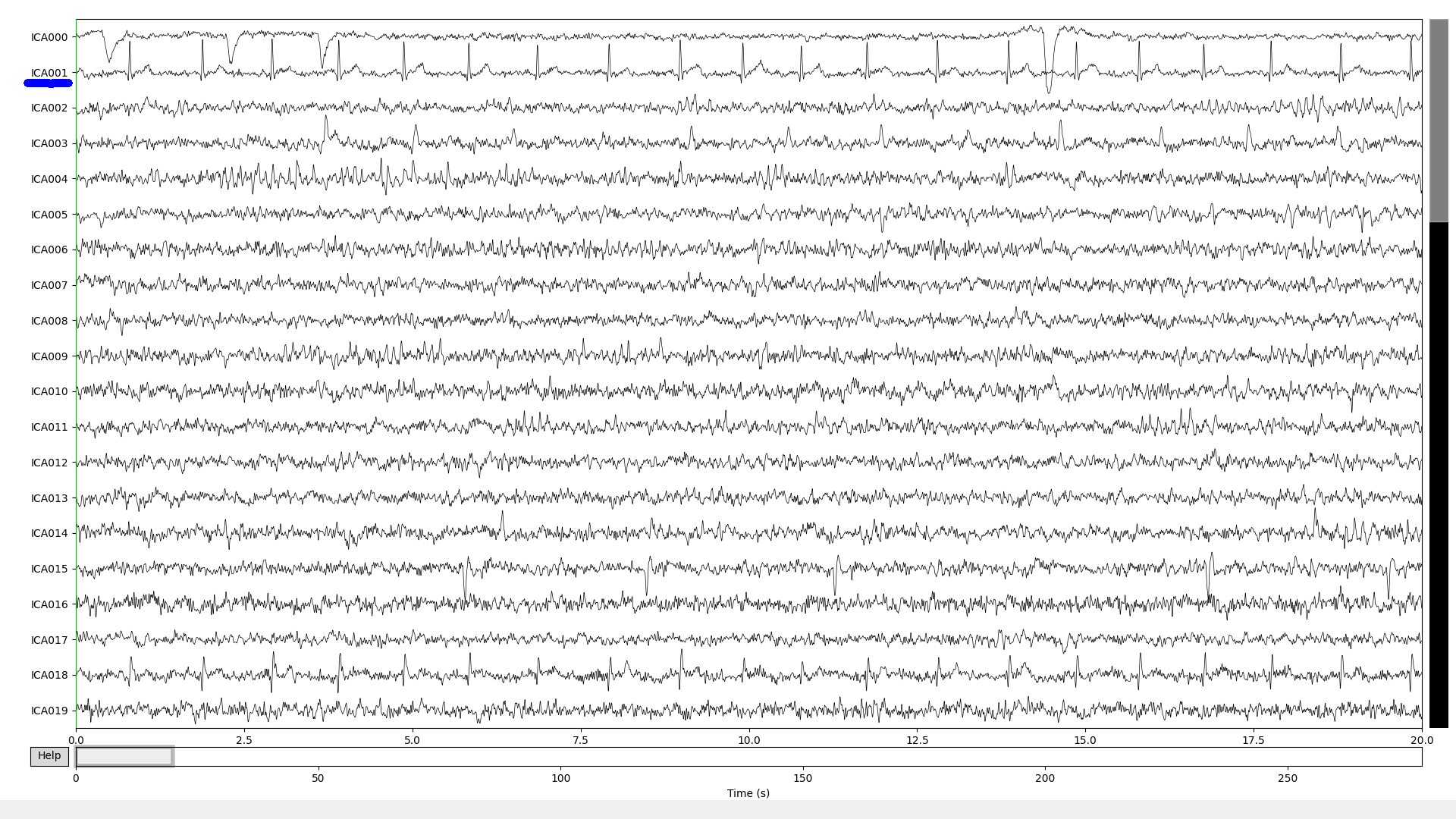
threshold=0.21Describe your solution
In the reference, the threshold was set to Pk >= 10^-20. Pk can be computed from formula (13) with λ and λ will change according to the number of data points. Thus, the threshold will also change according to the sampling rate.
The threshold of the Kuiper index can be determined easily:
If that's reasonable, I can make a PR for this implementation.
Additional context
The test script: