@GuillaumeFavelier thoughts? any way to expose such options?
Closed kdoelling1919 closed 3 years ago
@GuillaumeFavelier thoughts? any way to expose such options?
Yeah, I prefer the result with transparency
any way to expose such options?
plot_alignment has so many parameters already, I would rather not add another one. I prefer the auto-detection approach. I think computing the bounds of the brain surface is enough as a compromise between precision and computational cost.
Currently we make the innermost surface opaque (alpha=1). It's probably fine instead just to always make the innermost surface translucent (alpha=0.75 or something). That way you can see stuff behind it anyway, which is nice. Would that work for your sEEG example?
These are the examples where plot_alignment is used:
https://mne.tools/dev/generated/mne.viz.plot_alignment.html#examples-using-mne-viz-plot-alignment
@kdoelling1919 if you want to make a pull request with this change, if you could make minor improvements to wording if possible (otherwise just add an extra PEP8-compatible newline somewhere) to some of them, it will cause CircleCI to render them so we can see the outputs. Ideally you could pick examples that cover the various uses of surfaces that we have. I'd guess there would be 4 or 5 different ways we use surfaces in our examples.
In a worst case we can make surfaces be a dict mapping surface names to alpha values, but I'd rather avoid that API change if possible.
That's wonderful! That's what I want!
OK. So i'm not totally clear where we've landed. Seems like options are:
I'm happy with any of them. Though I can imagine that if we changed everything to .75 (option 2), some one else will be writing an issue soon enough complaining that they really NEED it to be fully opaque. so I'm inclined to make a more specific change like option 1 or 3.
Once there is consensus, I'll make the PR along with the tutorial changes as @larsoner suggested.
Though I can imagine that if we changed everything to .75 (option 2), some one else will be writing an issue soon enough complaining that they really NEED it to be fully opaque.
It's possible but really I think the transparent was probably always a better choice. We make aesthetic/functional judgments like this often for our plotting functions. This is why I wanted to make sure that, in your PR, you make some tiny change to each plot_alignment surfaces use "type" so that we can verify things got better in them, not worse.
So I say we try (2) and look at the results. If they look bad, I'd prefer (3) to (1). Making it contingent on sEEG/ECoG does not solve the problem -- for example if you plot EEG+scalp and your dig point is off by a couple of mm it will be inside. In that case should the scalp be transparent? So now the transparency of the surface depends on the locations of the electrodes, etc. It's cleaner to have a standard behavior in all electrode + surface combinations if possible.
@BarryLiu-97 by adding surfaces=["pial", "head"] or just surfaces=["pial"] (for only brain and no skull) as a parameter to plot_alignment should look like this
plot_alignment(raw.info, subject=subject, subjects_dir=subjects_dir,
trans=trans, show_axes=True, surfaces=["pial", "head"])o
I agree with you. Why not set this para Customizable? So people can choose whether transparent or not?
@BarryLiu-97 by adding
surfaces=["pial", "head"]or justsurfaces=["pial"](for only brain and no skull) as a parameter toplot_alignmentshould look like thisplot_alignment(raw.info, subject=subject, subjects_dir=subjects_dir, trans=trans, show_axes=True, surfaces=["pial", "head"])
Thank you so much!
I'm skeptical that option 2 will work, but I'm willing to be proven wrong.
I routinely use plot_alignment to check my coreg results by plotting scalp surfaces and dig points. I know from various problems in mne_coreg that being able to see the dig and FID points on both sides of the head through the scalp surface is not helpful and is generally confusing. Hence my concern.
Like i said I'm happy to be proven wrong but please consider use cases like https://mne.tools/stable/auto_examples/visualization/plot_eeg_on_scalp.html#sphx-glr-auto-examples-visualization-plot-eeg-on-scalp-py when evaluating the change.
Thanks Luke
On Wed, Oct 28, 2020 at 9:54 AM Keith Doelling notifications@github.com wrote:
@BarryLiu-97 https://github.com/BarryLiu-97 by adding surfaces=["pial", "head"] or just surfaces=["pial"] (for only brain and no skull) as a parameter to plot_alignment should look like this
plot_alignment(raw.info, subject=subject, subjects_dir=subjects_dir, trans=trans, show_axes=True, surfaces=["pial", "head"])
— You are receiving this because you are subscribed to this thread. Reply to this email directly, view it on GitHub https://github.com/mne-tools/mne-python/issues/8445#issuecomment-717949040, or unsubscribe https://github.com/notifications/unsubscribe-auth/ABKTXHOUH4CQYXVAYHAHGRTSNAPCPANCNFSM4TCHJWIA .
Like i said I'm happy to be proven wrong but please consider use cases like
I'm thinking/hoping that this example will look better with alpha=0.75 because you'll get more information -- you'll see the 50%+ of the electrodes not currently visible from that view.
So perhaps we should take a hybrid approach -- implement 2 and 3, so that in the (hopefully rare) cases where things do look worse, people have a way to get back to the old behavior. This is the approach we've been trying to take with topomap plotting and it has seemed to work well so far.
Concretely @kdoelling1919 I would implement this by:
plot_alignment with surfaces (I'd expect this to be 4 or 5 examples)0.75 is actually better (in the vast majority of cases) or not.surfaces=dict(...) support.Does that sound like a reasonable plan?
Why not set this para Customizable? So people can choose whether transparent or not?
FYI @BarryLiu-97 the reason I was trying to avoid this is that every time you add more options for people it increases code complexity and maintenance burden at our end, and also makes it harder for people to understand (more options = more complicated interface). But I agree it's probably safer to allow it...
Why not set this para Customizable? So people can choose whether transparent or not?
FYI @BarryLiu-97 the reason I was trying to avoid this is that every time you add more options for people it increases code complexity and maintenance burden at our end, and also makes it harder for people to understand (more options = more complicated interface). But I agree it's probably safer to allow it...
I couldn't agree more.
I'm thinking/hoping that this example will look better with alpha=0.75 because you'll get more information
here is the output of mne coreg with 0.75 head opacity..
mne coreg -s fsaverage -f ~/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif -d ~/mne_data/MNE-sample-data/subjects/ --head-opacity=0.75

I don't think that additional information is helpful in this case. Which points are inside the head vs on the opposing side of the head?
I don't think that additional information is helpful in this case. Which points are inside the head vs on the opposing side of the head?
Yes I agree this looks confusing in this specific case.
Yes I'm in agreement with @bloyl. Running these tutorials locally, I found that just making everything by default more transparent will make some other use cases harder to read. The reason I liked using only sEEG as a special case for transparency is because it's the only case we have (that I can think of) where you would purposefully have sensors inside the surface of interest, so I think it really is a special case that should be treated differently. Even in the scenario @larsoner mentioned, where one digitized point is accidentally inside the head, I'm not sure I would catch it from a plot like @bloyl just presented.
Yes I'm in agreement with @bloyl. Running these tutorials locally, I found that just making everything by default more transparent will make some other use cases harder to read. The reason I liked using only sEEG as a special case for transparency is because it's the only case we have (that I can think of) where you would purposefully have sensors inside the surface of interest, so I think it really is a special case that should be treated differently. Even in the scenario @larsoner mentioned, where one digitized point is accidentally inside the head, I'm not sure I would catch it from a plot like @bloyl just presented.
I found a para seeg(bool) in plot_alignment, what if set alpha=0.75 when seeg=True?
here is the output of mne coreg with 0.75 head opacity..
I think you get this because of your (unfortunately persistent) graphics problems. It looks better with a modern graphics setup / advanced rendering. But we still do block the points on the far side (that are not inside the head) intentionally in that case actually.
Running these tutorials locally, I found that just making everything by default more transparent will make some other use cases harder to read.
It turns out that hacking in a head_alpha=0.75 instead of 1.0 in plot_alignment is not enough -- you need to also enable depth peeling (we'll need to talk to @GuillaumeFavelier about enabling this properly; for now you can test by changing this line to True) this plot:

Becomes this:

To me this is nicer, but probably still not good enough for sEEG. And adding depth peeling things become more challenging (and restrictive in terms of which graphics cards actually work). So I can live with leaving the default at 1.0 and just having surfaces=dict support.
I found a para seeg(bool) in plot_alignment, what if set aplha=0.75 when seeg=True?
My proposal is to make it so you can do surfaces=dict(head=0.1, dict(pial=0.5) or whatever you want to set the surface alpha(s).
Here's what sEEG looks like with depth peeling enabled. I would be fine with this. Particularly since the skull is unnecessary anyway.
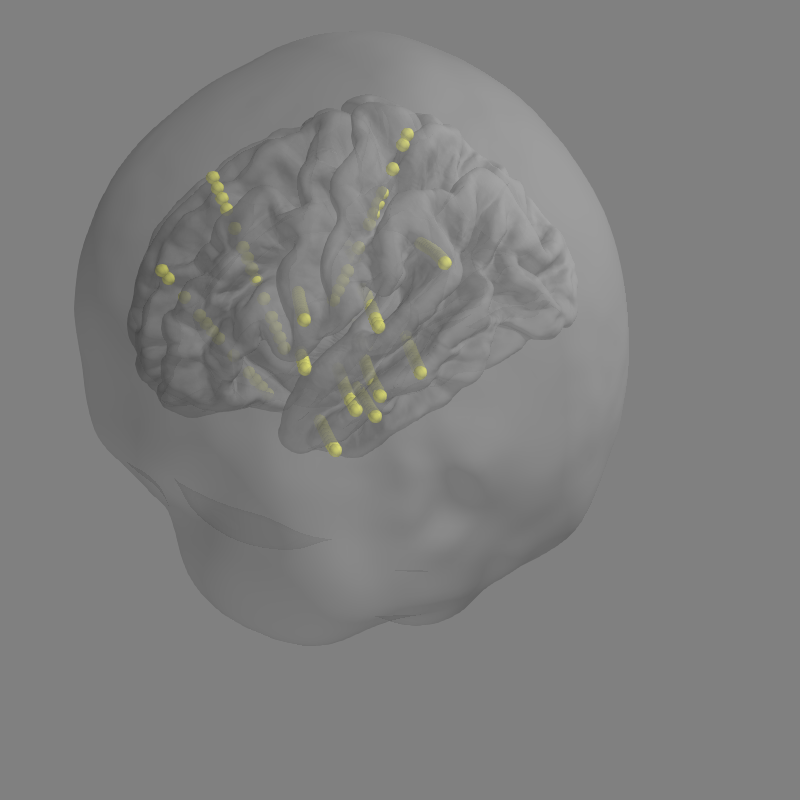
Alternatively, if we do go with adding surfaces=dict(...) functionality. I would still recommend that the default be some transparency when seeg electrodes are involved. Using dict for alpha settings will already be unwieldy so I'd rather the defaults be as sensible to the case as possible.
I would still recommend that the default be some transparency when seeg electrodes are involved.
Fair enough. I think this line:
alphas = (4 - np.arange(len(skull) + 1)) * (0.5 / 4.)Is just a really convoluted np.linspace call of some sort. I would first convert this to the equivalent np.linspace call, then set the upper bound to 1. or 0.75 (or whatever looks good) if seeg electrodes are pleasant.
I like the last figures @larsoner and @kdoelling1919 produced.
This issue is fixed by #8446
Describe the new feature or enhancement
Using plot_alignment to see where sEEG electrodes are located in the brain. But if you you use any brain surfaces such as
surfaces=["pial"]the brain is fully opaque so you can't see where the electrodes are that are inside the brain. So it would be best to give the brain a bit of transparency when you have sEEG electrodes embedded in the brain.Describe your proposed implementation
As far as I can tell, this is a single line change. I have already created a feature branch in my fork which changes the setting of opacity depending on whether there are sEEG electrodes present for plotting. https://github.com/kdoelling1919/mne-python/blob/5b039cdfb3da64c89947f09a0bfd83481955f29c/mne/viz/_3d.py#L840
Describe possible alternatives
Another option might be to set transparency generally any time there are sensors to be plotted that are inside the brain volume. Sounds like a bit more work (and more knowledge about the surfaces than I currently have) but it would be more general.
Additional comments
Here is what the plot_alignment looks like in current master version of plot_alignment
Here is what it looks like in my branch.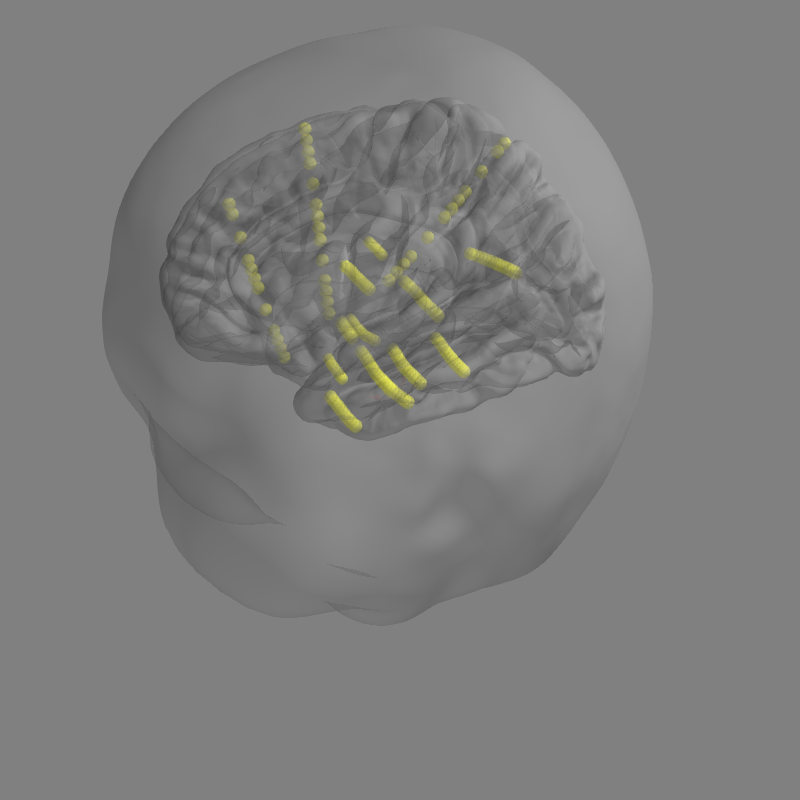
If people agree this is useful beyond just for myself. I'll make the pull request.
Thank you!