I think what looks incorrect to me is that you have:
a, b, diag = cca(x, y, backend) # a.size() = [D1, C]
a, _ = torch.linalg.qr(a) # a.size() = [D1, C]
alpha = (x @ a).abs_() # alpha.size() = [B, C]but you need to compute x_tilde first:
a, b, diag = cca(x, y, backend) # a.size() = [D1, C]
x_tilde = x @ a # x_tilde.size() = [B, C]
x_tilde, _ = torch.linalg.qr(x_tilde) # x_tilde.size() = [B, C]
alpha = (x_tilde.T @ a).abs_() # alpha.size() = [C, D1]in one line the difference btw the two is more obvious (ignore the extra orthonormalize for clarity):
a, b, diag = cca(x, y, backend) # a.size() = [D1, C]
alpha = ((x @ a).T @ a).abs_() # alpha.size() = [C, D1]it seems that is different from what anatome currently has?
Why does anatome compute pwcca by orthonormalizing the
avector instead of the CCA vectorx_tilde = x @ a? e.g.https://github.com/moskomule/anatome/blob/393b36df77631590be7f4d23bff5436fa392dc0e/anatome/distance.py#L161
the authors do the latter i.e. orthonormalize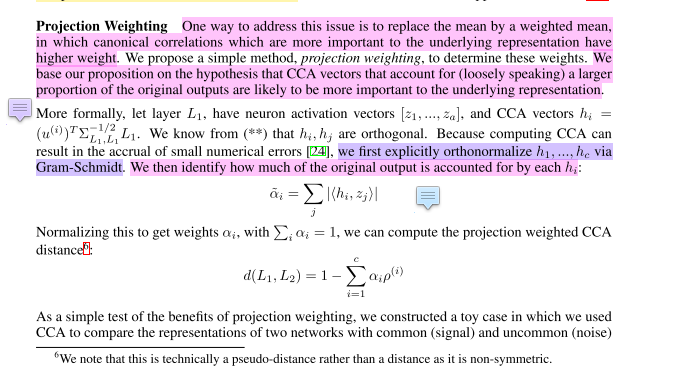
x_tildenota:https://arxiv.org/abs/1806.05759
current anatome code:
related: https://stackoverflow.com/questions/69993768/how-does-one-implement-pwcca-in-pytorch-match-the-original-pwcca-implemented-in