@royerloic how do you feel this functionality intersects with the current Path object in the shapes layer? For example see this WIP PR #532 for examples of lines in 3D? You can pass a list of NxD arrays as input and a list of colors and it will do 2D and 3D rendering. It won't change transparency along the line, this could be done, but unclear if it is needed functionality. There's also only one global width for each line right now.



🚀 Feature
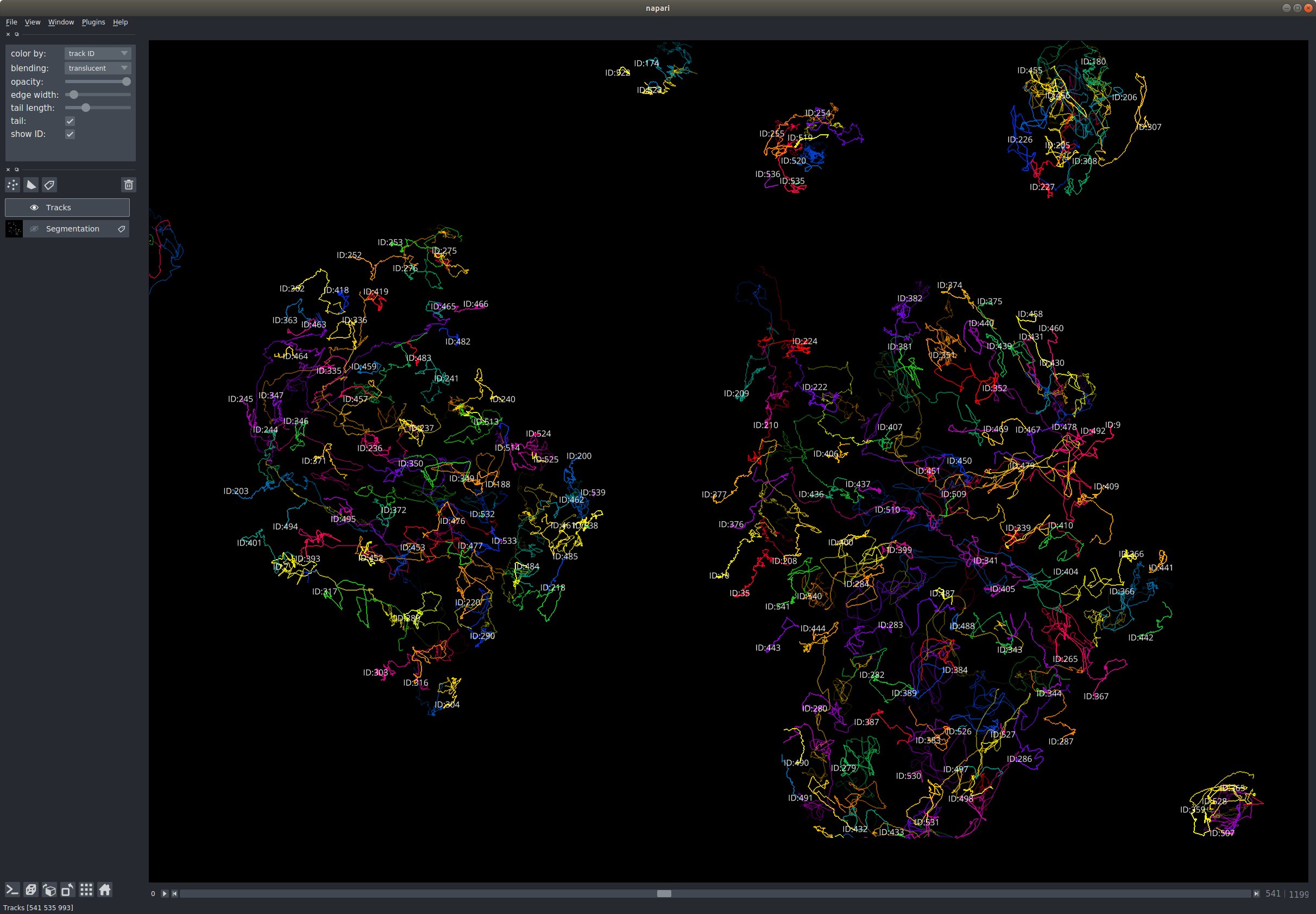
A typical bioimage analysis task is to track the movement of objects of interest such as cells in their 2D or 3D movement (possibly more, @jni --guardian-of-dimensions-- would argue). Having the possibility to display such tracks both in the 2D and 3D view and show future and past positions (increasingly faint and transparent as you go away from the current time point) would be extremely useful. The possibility to edit such tracks and adjust the positions of objects would be cool too to refine the output of automated algorithms. Dealing with object division can be done by ending a track and restarting two tracks. Basic styling of the thickness, opacity of the tracks, and how far back they extend in the past or future, would be great too.
Tracks could be represented as a collection of n*k arrays where n is the length of the track, and k is the dimension of the space occupied by the objects.