Thanks Gytis ⭐️ An auspice implementation would be desirable as it would allow us to group subtrees interactively based on any non-continuous trait which is set on internal branches (i.e. augur traits has been run in the preparation of the dataset). Implementation wise, this seems simplest via (a) restricting it to rectangular layouts, (b) marking state-change branches as hidden, (c) writing a novel y-value setter for the tree (but of course the devil is in the details!). Importantly this wouldn't require modifying the underlying nodes structure in auspice.
Context
Phylogenetic trees are inherently difficult to interpret. Whilst colouring the tree by inferred ancestral state (geographic location, host, resistance, etc) depicts the full history of state changes for every lineage across the tree it is nonetheless not easy to interpret, especially for non-experts. By showing migrations on a map the interpretation issue is ameliorated but only addresses the number of state changes and locations involved, leaving the user to interpret the fate of lineages in the new state from the tree by themselves.
By splitting and grouping lineages that have been introduced into a new location for as long as they stay in the same location one can get a sense of how many times lineages have been introduced and how well they did in the new location. Doing so gives a very visual interpretation of how suitable locations are for local transmission and intensity of migration over time. Locations that are poorly connected but suitable for local transmission would be indicated by few but large subtrees, locations that are well connected but unsuitable for local transmission as many small subtrees and so on.
Description
Examples
Martha Nelson's work is the first example where I have found this type of visualisation: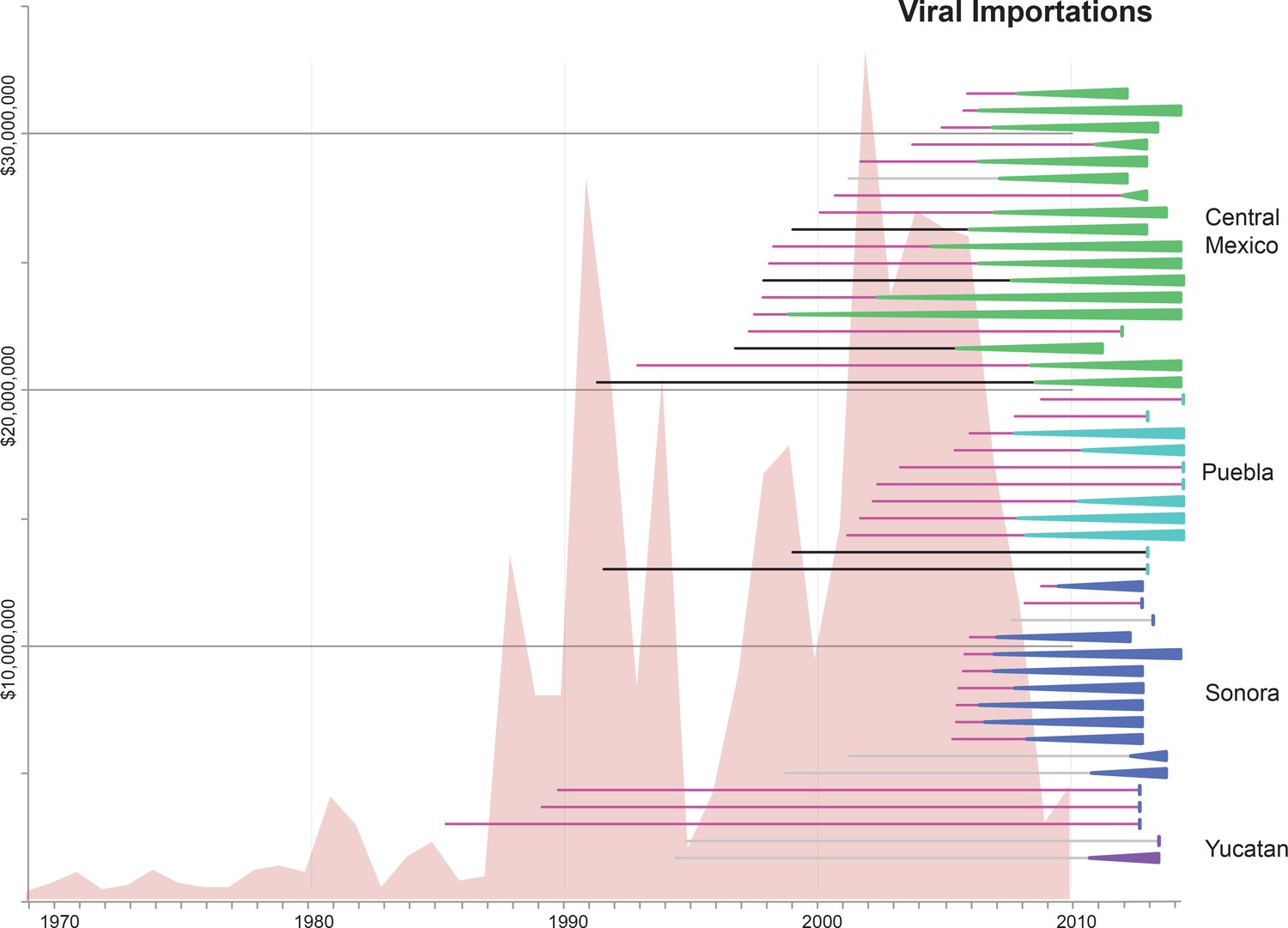
Here's a more extensive Ebola example though. This tree contains all of the information about the migration history of Ebola virus in West Africa during the epidemic in 2014-2016 but is not easy to interpret (even with a better aspect ratio).
However, breaking apart the tree using the method described above at the country level shows an easily interpretable picture: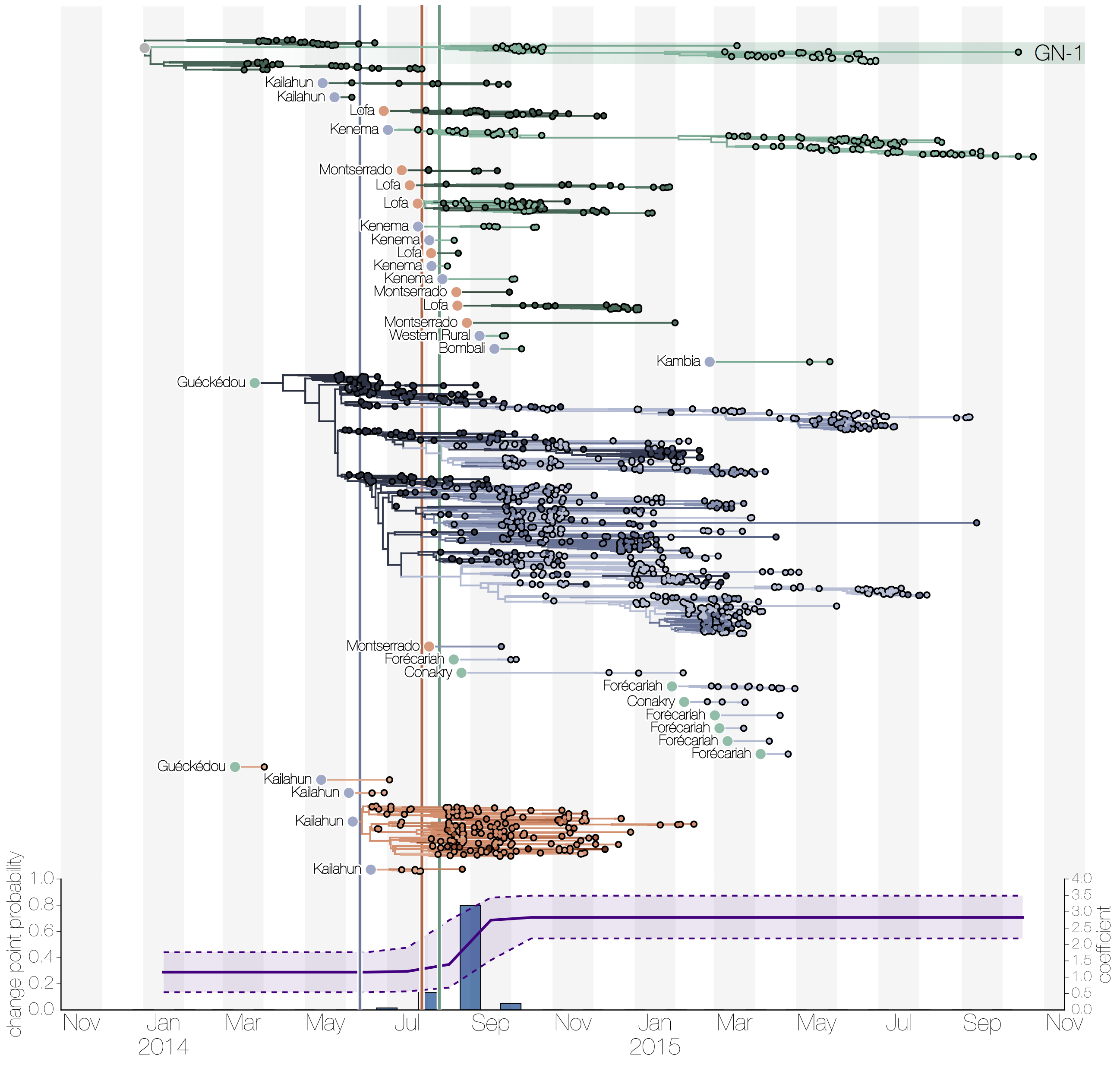
Subtrees inferred to be in Guinea (green) are small but there's many of them, suggesting frequent introductions without much amplification of viral lineages via local transmission. Introductions originate (indicated by circle colour at the base of each subtree) from both Sierra Leone (blue) and Liberia (red). There are fewer subtrees in Sierra Leone (blue) but one of them is enormous, indicating that introductions were rarer (and primarily from Guinea) but that local transmission in Sierra Leone was rampant, leading to a local outbreak that is largely dominated by the one single lineage. Liberia (red) sits somewhere in the middle - its outbreak too is dominated by a single introduction but it is smaller and shorter with a few small introductions.
The splitting and grouping of subtrees visually explains the case data that we see perfectly - Guinea had a low level but protracted outbreak because Ebola virus was not able to spread as efficiently and instead cases were primarily generated via multiple introductions. Sierra Leone had a very intense peak with ~700 new cases reported per week, fuelled almost entirely by local transmission with Liberia being similar but slightly lower. I don't have a figure for number of cases but you can see everything together in this video.
Repeating the same process of splitting and grouping subtrees at a smaller level (administrative division level rather than country level) was even more illuminating though not looking quite as good. Even though we saw how the virus population was distinctly distributed between the three countries no such distinct clustering exists within each of the countries (with maybe the exception of Liberia). Importantly this answers a question many had at the time of the epidemic which is what role urban centres were playing in the region - were they persistently infected and seeding outbreaks elsewhere in the country or were the outbreaks short-lived everywhere. It was the latter.
Another example is MERS coronavirus. This is a viral infection that has been jumping from camels into humans since 2012 but the virus unlike SARS-CoV-2 doesn't transmit between humans well. Once can look at evolutionary history of MERS-CoV infections in humans (blue) and camels (orange, obviously) jointly: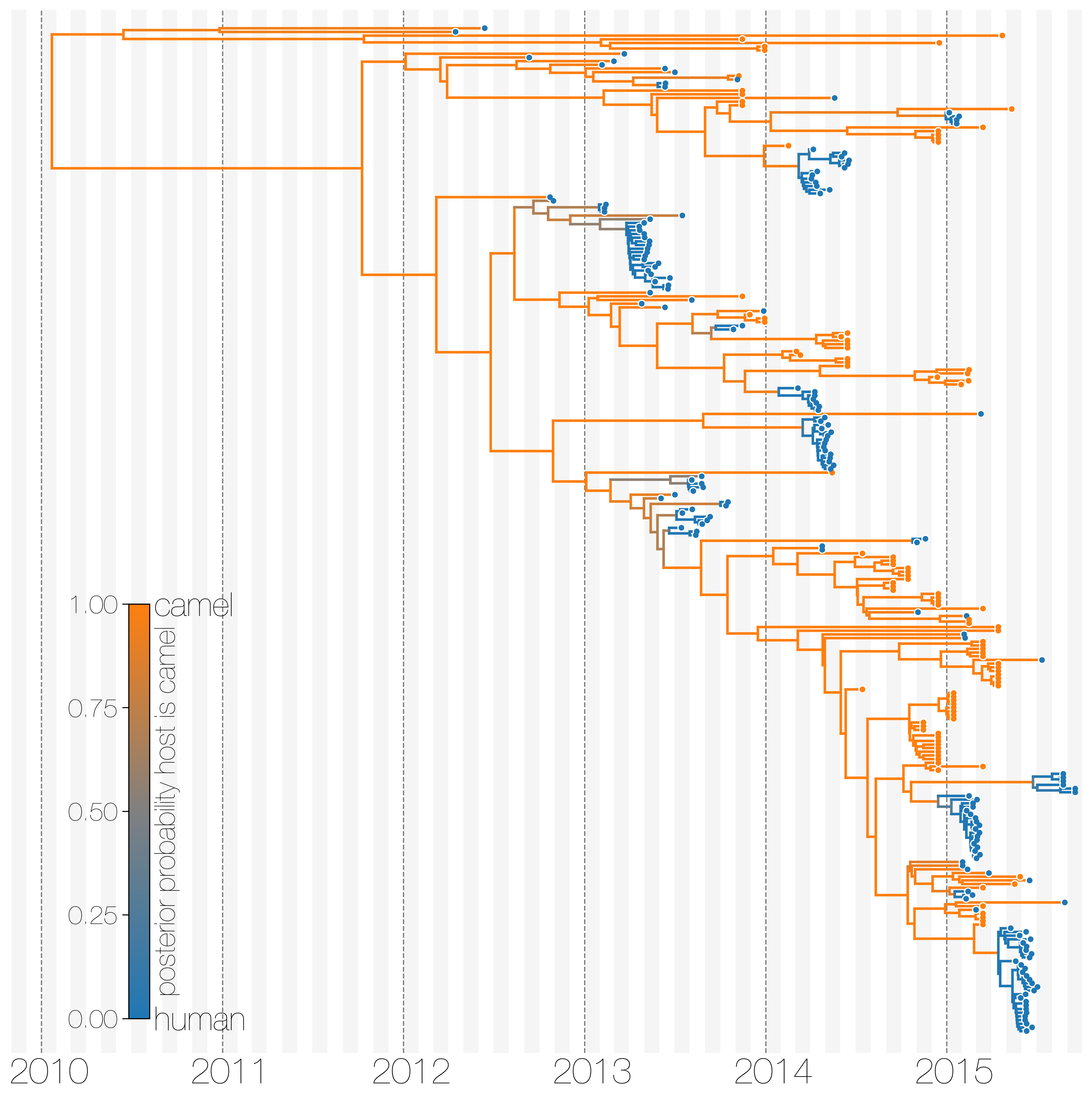
or one can split and group lineages based on host change and see clearly that cross-species transmissions are only camel-to-human and very unsuccessful in humans:
Possible solution
One solution is to recompute y axis coordinates for every subtree such that they end up close to each other based on location and then hide their parental branches so there's no criss-crossing of vertical lines across subtrees.
Another has been discussed previously by Trevor and James before. Since auspice JSONs cannot contain multiple trees the most straightforward solution is to produce a JSON that contains all of the subtrees as descendants of the root node with the branch leading from the root to the base of the subtree invisible (I think this is possible). This might need to happen at augur level.