If I create a sphere with eg:
view = nglview.NGLWidget()
view.shape.add_sphere([0.0, 0.0, 0.0], [0,0,1], 10.0)is there a way of setting the opacity of the sphere?
Thanks in advance!
Closed hainm closed 4 years ago
If I create a sphere with eg:
view = nglview.NGLWidget()
view.shape.add_sphere([0.0, 0.0, 0.0], [0,0,1], 10.0)is there a way of setting the opacity of the sphere?
Thanks in advance!
ah, there's actually an API for it but you have to play with it with more complicated case (mixing structure, shape, ...).
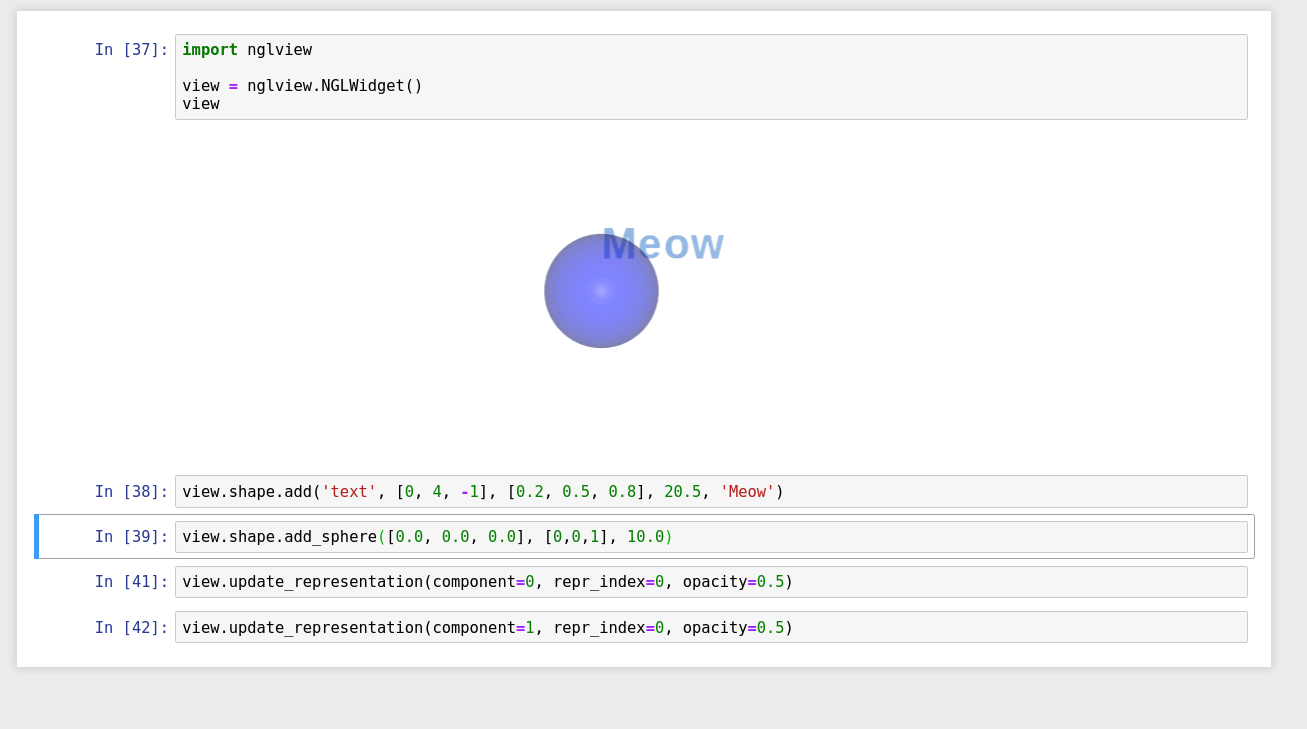
ah, there's actually an API for it but you have to play with it with more complicated case (mixing structure, shape, ...).
Great, thanks!
I'm trying to figure out the best way to set the orientation. Here is my current procedure. It works. However, can you let me know if this is indeed the best way. The reason that I wasn't sure is that it uses two private methods:
Manually set the orientation to whatever I want by rotating the object.
Get that orientation (as a 16-number list) with:
mat = view._camera_orientationIn the future, set that orientation with:
view._set_camera_orientation(mat)If I want to make sure it is also centered, call:
view.center()Hi,
It’s currently the best way. Cheers
Hai
On Sun, Apr 28, 2019 at 3:38 PM Jesse Bloom notifications@github.com wrote:
I'm trying to figure out the best way to set the orientation. Here is my current procedure. It works. However, can you let me know if this is indeed the best way. The reason that I wasn't sure is that it uses two private methods:
1.
Manually set the orientation to whatever I want by rotating the object. 2.
Get that orientation (as a 16-number list) with:
mat = view._camera_orientation3.
In the future, set that orientation with:
view._set_camera_orientation(mat)4.
If I want to make sure it is also centered, call:
view.center()— You are receiving this because you authored the thread. Reply to this email directly, view it on GitHub https://github.com/arose/nglview/issues/785#issuecomment-487409212, or mute the thread https://github.com/notifications/unsubscribe-auth/ABB645J4BKIFR75FYLHPWUDPSX4LNANCNFSM4HHSP2TA .
view._set_camera_orientation(mat)
@jbloom I've checked and we should use view.control.orient(...) function.
view.control.orient(mat) # from view._camera_orientationHi everyone, I am using gro and xtc files from gromacs and visualize them using nglview within Jupyter notebook. It is unclear why rendered and downloaded images from nglview package contain white slits as you may be able to notice them on the second picture. It's not too big of a problem. Thank you for your help in advance!
The version of packages is as follows: nglview version = 1.1.9 mdanalysis version = 0.19.2
I also had @mktonycho's problem with vertical lines, although they go away if I use antialias=True with render_image.
Hi
Yeah, that is the current solution. I will open an issue in NGL. Thanks.
On Tue, May 14, 2019 at 8:43 AM Jesse Bloom notifications@github.com wrote:
I also had @mktonycho https://github.com/mktonycho's problem with vertical lines, although they go away if I use antialias=True with render_image.
— You are receiving this because you authored the thread. Reply to this email directly, view it on GitHub https://github.com/arose/nglview/issues/785?email_source=notifications&email_token=ABB645JKB6UPMPLZ74SRYKDPVKXWFA5CNFSM4HHSP2TKYY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGODVLLFLY#issuecomment-492221103, or mute the thread https://github.com/notifications/unsubscribe-auth/ABB645KUL5HCGCSRUTVV3L3PVKXWFANCNFSM4HHSP2TA .
It is unclear why rendered and downloaded images from nglview package contain white slits as you may be able to notice them on the second picture. It's not too big of a problem.
@mktonycho should be fixed in master branch (by upgrading to NGL 0.36).
@jbloom @hainm Thank you so much for your help!!! I will try again with the new version!
Hi! Not sure if this should be submitted as a separate issue, or if there is something I'm not doing correctly that can easily be fixed on my side.
I'm trying to run nglview in a Jupyter Notebook through ASE using nglview.show_ase(atoms) where atoms is the ASE Atoms-object I want to view.
When I make this call, I get a permission error is thrown (pasted below).
Appreciate any help I can get here.
PermissionError Traceback (most recent call last)
hi @rasmusthog, you can do either
C:\cygwin64\tmp writable by pythonget_structure_string function to write file to your current folder.something like
def get_structure_string(self):
print("HELLO")
fname = 'tmp.pdb'
self._ase_atoms.write(fname)
with open(fname) as fh:
return fh.read()
nv.adaptor.ASEStructure.get_structure_string = get_structure_stringHi,
I'm trying to get nglview up and running and I'm probably overlooking something simple...
I created an environment with conda and installed juypter, pytraj and nglview. Everything installed smoothly. When I run the tutorial, I see the expected output from the first cell (nglview version = 1.1.7, pytraj version = 2.0.4)
However, after the third cell, (view._display_image()), I get a broken image icon. I tried running the same command sequence in ipython and got
Thanks for your help
@jaygn Please post your screenshot.
(But I guess you have to run render_image and _display_image in different cell, make sure there's a small delay between commands).
But you can try the latest version via (v2.1.0), you can see the rendered image without using _display_image.
Thanks @hainm. Here's the screenshot. I tried both with render_image and _display_image in the different and the same cells. I get the same behavior when I run the tutorial notebook in place or if I copy the commands to a new notebook and run it there.
I tried updating to 2.1.0 and I still don't see the image, although now it's trying to display after the render_image step as you said.

sorry for your inconvenience. Can you try
jupyter-nbextension install nglview --py --sys-prefix
jupyter-nbextension enable nglview --py --sys-prefixThat worked! It looks great now. Thanks so much for the prompt help!
thanks for trying.
Thanks, @hainm!
I have now modified the permissions (changed the ownership of the tmp-folder to my user instead of the local Administrator account), but I'm still getting the same error. Also tried changing the permissions so that the tmp-folder is globally readable (chmod -R 777), but still the same result.
This is how the folder looks from a ls -l:
drwxrwxrwx+ 1 [my username] [computer name]+None 0 May 30 10:27 tmp
sorry @rasmusthog that I can't not help further with the permission error because I don't have any windows machine to test (I am mainly working with linux and macos). But please try the patch I mentioned. cheers.
I'm embedding NGL. I'm loading a model like this (in an async typescript function):
let obj = await stage.loadFile(name, { defaultRepresentation: false })
obj.setDefaultAssembly('BU1')
representations.forEach((elt, idx) => {
obj.addRepresentation(elt[0] as NGLViewer.StructureRepresentationType, elt[1])
})
obj.autoView()but when I dump the Three.js scene (stage.viewer.scene) immediately after this, the graphics objects don't exist yet. I think they're getting created in the background. Is there any callback or hook so I can know when the world is fully loaded?
@garyo please see example(s) here:
https://github.com/arose/ngl/blob/master/examples/scripts/representation/distance.js
Thanks for the example -- but I don't see there how to invoke a callback once all the graphics objects are loaded. If you dump the scene at the end of that example, just after o.autoView(), it will not have any meshes. Actually in your simple example it may, but if you use a 'surface' representation or something else that requires a bg task to compute, it won't be there yet.
Looking at the source, in representation.ts representation.make() takes an optional callback to call when it's done, but nobody calls it with a callback arg as far as I can see.
@garyo I see. Do you mind opening the issue in NGL repo (we are in nglview repo now)? There are many experts there.
ok, done -- thanks
Hi,
I want to ask if I can display hydrogen bonds with nglview.
I thought the add_distance or add_contact method played that role but is it not?
Thanks for your help.
@yamasakih yes, you can use add_contact for that (v.add_contact(hydrogen_bond=True))
You can try the NGL website for different types of contacts (Choose "Examples" -> "representation/contact"). cheers.
It worked! Thanks so much for your prompt help!
Hi,
[EDIT] I have found view._set_size() works.
I tried to use view.layout.height = '600px' to adjust the view height but it doesn't work. The widget height is increased but the viewport(?) height is not changed. The width control works. Is there any other ways I can change the height?

Thanks!
Hi
Please use view._set_size for now. The issue will be fixed in next release.
Hi @hainm!
Finally got time to do the patching. It works fine now :) Thanks!
Has anyone used NGLview in a JUpyter notebook to browse through docking results? Protein as a PDB docking results as .sdf.gz
@drc007: do you have a .sdf.gz file? Have you tried yet or you don’t know how?
Hi @hainm, I've displayed a pdb file but I don't know how, or if it is possible to browse through a file of docked structures superimposed on the protein.
I've written the Jupyter notebook to do the docking and have the output. I could use an external program to view the results but was wondering if it was possible to view them in the notebook?
I've displayed a pdb file but I don't know how
I am not sure what you meant here. Have you tried “nglview.show_file(“your.pdb”)?
Hi! I would like to know if is possible to add a colorbar based on b-factor values of a protein.
Hi
Coincidently I also have a question about b-factor colouring.
I have some data values stored in the b-factor column of a PDB file that range from -1 to 1. I would like to colour the atoms according to these values with -1 = red, 0 = white and 1 = blue, with smooth red-white and white-blue gradients for intermediate negative and positive values. Is it possible to specify this in nglview?
I've displayed a pdb file but I don't know how I am not sure what you meant here. Have you tried “nglview.show_file(“your.pdb”)?
I can display the pdb fine, I was wondering if anyone had ever developed a notebook where you could display the pdb, then scroll through a file containing docked structures and display the docked structure in the protein?
I would like to know if is possible to add a colorbar based on b-factor values of a protein.
I have some data values stored in the b-factor column of a PDB file that range from -1 to 1. I would like to colour the atoms according to these values with -1 = red, 0 = white and 1 = blue, with smooth red-white and white-blue gradients for intermediate negative and positive values. Is it possible to specify this in nglview?
@OCientista and @njbruce:
There is no direct way to do those in nglview, but you can check the custom color scheme from https://github.com/jbloomlab/dms_struct/tree/master/dms_struct (@jbloom).
ping @arose in case you have any idea.
Hi,
Is it possible to get a list of all the interactions once I added the contact representation? Thanks for the amazing work!
Tony
hi @tonyyzy: unfortunately no. cc @arose in case you know there's any.
hi @tonyyzy: unfortunately no.
@hainm I know there is contactStore from NGL, is there a way I can poke into javascript from NGLview?
glad that you asked. Yes, you can hack it (and let me know if you can pull the data). Can you try?
# 1st cell
view._execute_js_code("""
this.send({"type": "data", "data": "your data here"})
""")
# 2nd cell (give sufficient time for data traveling from JS to python)
print(view._ngl_msg)ah, you can access NGL.Stage via this.stage
@hainm Thanks a lot!
Took some effort, here it is
# 1st cell
view = nv.show_pytraj(traj[0:1])
view.clear()
view.add_ball_and_stick("ligand")
view.add_cartoon()
view.add_contact()
view
# 2nd cell
view._execute_js_code("""
this.send({"type": 'data', "data": JSON.stringify(this.stage.compList[0].reprList[2].repr.bufferList[0].picking.contacts.contactStore.index1)})
""")
# 3rd cell
view._ngl_msg
Output: {'type': 'data',
'data': '{"0":0,"1":0,"2":1,"3":3,"4":3,"5":4,"6":6,"7":6,"8":8,"9":9,"10":9,"11":10,"12":11,"13":12,"14":13,"15":15,"16":15,"17":16,"18":16,"19":17,"20":18,"21":23,"22":26,"23":26,"24":27,"25":28,"26"...
However, _ngl_msg attribute needs to be updated hence the 2nd and 3rd cell cannot be merged into one. I tried time.sleep which blocked the main thread for update; threading.Timer execute the next command directly. Any idea how to work around this? I want to loop through 2000 frames and determine all the interactions of each frame...
@tonyyzy you can create a callback function to handle message from JS whenever the message is ready.
data = []
def handle_contact(self, msg, _):
if msg['type'] == 'contact_data':
data.append(msg['data'])
view.on_msg(handle_contact)# 2nd cell
view._execute_js_code("""
this.send({"type": 'contact_data', "data": JSON.stringify(this.stage.compList[0].reprList[2].repr.bufferList[0].picking.contacts.contactStore.index1)})
""")NOTE: I change "data" keyword to "contact_data" from your example.
@hainm That's really neat! The only problem is that in a loop, the callback function doesn't have enough "time" to be executed if a lot of js commands are issued. So I had to set an async sleep and play with the sleep time a bit to make sure data from all frames are recorded :)
For any questions relating to how/why/ ... Or anything you don't know where to ask. For bug, suggestion, ... please open seperate issue
PS: QA-0: https://github.com/arose/nglview/issues/589