Just for sanity-checking I deleted all cache-folders on both the Linux-Server and my Windows 10 PC and reran the script one more time. Interestingly, now I also got the beta-images on my PC (and they seem to be the same as on the server), but still with respect to the SpecifySPMModel node on the server I have files with proper size (1,5GB) on Windows still two files with 0KB
Summary
This issue is possibly related to #3301. All problems discussed there were bypassed by tweaking the script so that it doesn't produce the error anymore (specifically, it was discussed that
SpecifySPMModelcannot handlebids_event_files). The following script bypasses this by usingsubject_infoinstead ofbids_event_files. The script now works when running on a Linux-Server via Singularity but not when using Docker + WSL2 on Windows 10 (files are saved on the local hard drive, not in the WSL-2 distro (Ubuntu 18.04 in my case), maybe this causes the problem?).Actual behavior
On my windows PC: The scripts runs through 'successfully' without reporting any errors. However, when it reaches the
SpecifySPMModelnode, this folder contains files of no size (0KB). These files of no size apparently still seem get passed over to to the next nodeEstimateModelwhich also runs through without reporting any errors but outputs no beta-images.On Linux: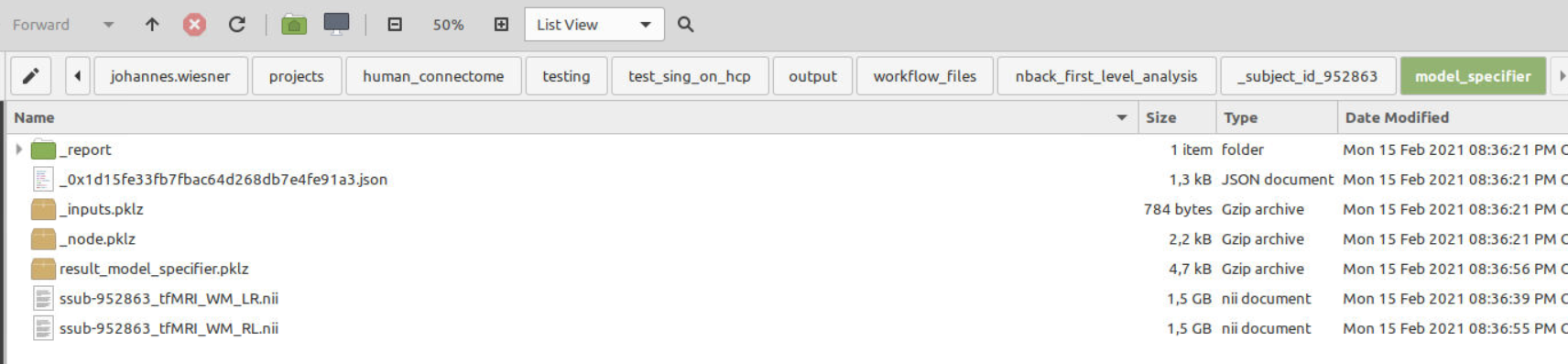
On Windows: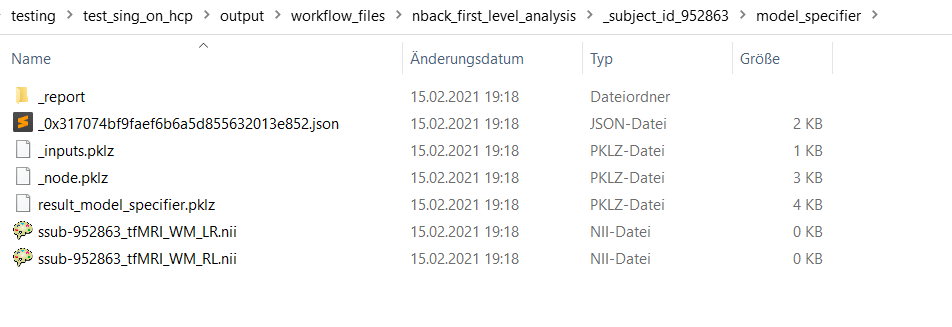
Script/Workflow details
This is the now tweaked code that works on the Linux-Server but not on my personal Windows 10 machine:
Platform details:
Same setup as in #3301:
Execution environment
Same setup as in #3301
Using Michael Notter's
nipype_tutorial(most-recent version miykael/nipype_tutorial:2020) running as a docker container on Windows 10 (+ WSL2, Ubuntu 18.04)