In MED (also gmsh in fact) we can give the same name to different sets corresponding to different manifold dimensions (so Top can be used to group all 2d elements on the top, and some 1d elements on the top), internally MED will just assign another unique set id.

So this motivates me to use the unique element set id's as keys in the mesh.tags (aka your field_data).
Reading the gmsh-converted .msh file containing two different sets with a same name leads to one being eaten by the other. So maybe we also need to redesign your field_data?

Hi all,
I wish to convert a mesh from SALOME (MED 4.0) to .msh or .xml alongside with cell attributes.
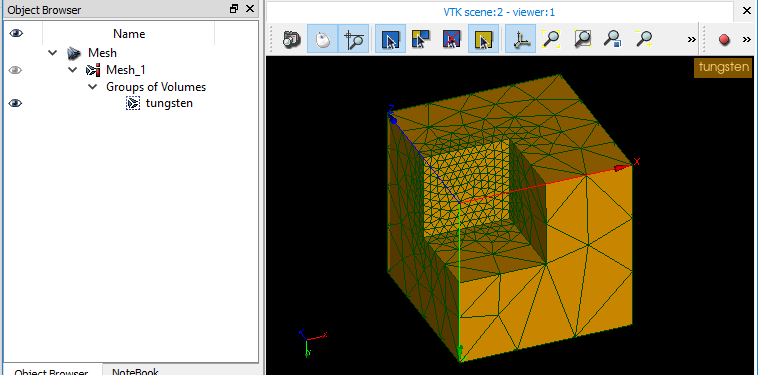
Here's an exemple of a mesh from SALOME with 1 volume marked. MED.zip
Meshio seems to read it without issues. But, when converting to .msh and after opening it in GMSH, here's the mesh file I get. gmsh.zip
No sign of the entity "tungsten".
Thanks guys for your help!
Rem
PS: Screenshot of the med file re-imported in SALOME, the group is there as expected.