General comments from the presentation
You showed the result of your research by using a subset of the input citations. Would it be possible to have a complete coverage of the results by considering all the citations, instead of just a part of it?
Yes, the results have been recalculated considering the entire dataset of input citations you provided. The new version of the output dataset was published with the following DOI: 10.5281/zenodo.4892551.
It would be great to compare the results you obtained if you use only the procedure (i.e. regular expressions) described in Xu et al. vs all those that you obtained by using your updated methodology, i.e. that one which includes more regex as you have presented.
It was interesting indeed. The procedure of Xu et al. was reproduced and their results compared with ours. The data were reported and interpreted in the new version of the article and the visualization has been updated to take into account this comparison. The visualization can be seen at the following address: https://open-sci.github.io/2020-2021-grasshoppers-code/.
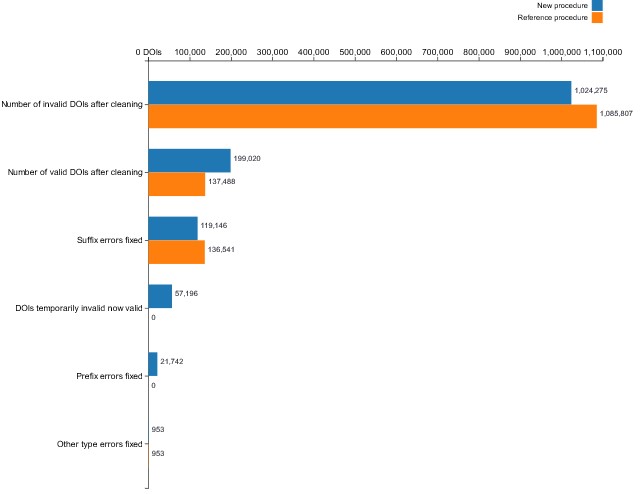
The comparison to the Xu et al. reference study showed that the methods used in this research could be employed to clean more DOIs overall. However, which could be seen as surprising, applying the methods from Xu et al. allowed 17,395 more suffix errors to be cleaned. On the other hand, the reference procedure did not clean any prefix errors in the given dataset. This result can be explained by taking a closer look at the regular expressions and the definition of prefix- and suffix-type errors used by Xu et al. In the study presented here, the regular expression for prefix errors was broader, as any additional strings before the DOI were considered prefix errors, even if they were not at the beginning of the string. Meanwhile, the reference procedure considered prefix errors only those at the beginning of the string, hence the absence of matches in the given dataset. The current study, for instance, in the DOI "10.1016/J.JLUMIN.2004.10.018.HTTP://DX.DOI.ORG/10.1016/J.JLUMIN.2004.10.018", considered the presence of the DOI proxy server as a prefix-error, because it appeared before the DOI. The higher number of suffix errors cleaned with the methods by Xu et al. comes as a consequence. Since the present algorithm subsequently checks a DOI name for prefix, suffix and, finally, other-type errors, the errors not caught as prefix errors are cleaned as suffix errors in the next step. Presumably, the mistake related to the presence of the DOI proxy server was fixed as a suffix error in the Xu et al. procedure, which led to a higher number of matches.
You said that it is pretty impossible that the process stop to compute the various values. But what does it happen if someone by accident switch off your computer while running the computation? Do you have a way to restart it from the last data produced, avoiding to start it again from the beginning?
Indeed, it is true, if the PC were to shut down, the entire work (except for the cache of requests to the DOI Proxy server) would have been lost. To deal with this problem, a new support method was implemented, read_cache. If a cache file was created, this support method reads the data processed up to that moment and restarts the process from the last CSV line read and not from the beginning. This function has been integrated into both check_dois_validity and procedure methods, that create or update a cache file every N DOI. The user can customize the number of DOIs after which the cache updates, as well as the location of the cache file. The code was inspired by the strategy adopted by the Leftovers 2.0 team to solve the same problem.
An invited expert identified a bug in the code for cleaning the DOIs. Is it possible that this bug affected the results presented? If so, it would be necessary to recompute the whole data, of course.
Yes, the results related to the application of regular expressions had to be recalculated and they were. However, it was not necessary to recalculate the results from the verification of the previous validity of the DOIs and they have been reused to speed up the process.
Did you check, in some way, if the cleaning of a DOI produced a valid DOI which is indeed the article that is indeed cited by the citing entity? In other words, is it possible that your cleaning process produce a citation between two entities which is not the one that was originally defined in the citing paper?
To check if the cleaned DOI names were part of the citing entity references, 100 DOIs were randomly selected from the results and manually checked. Three cases were differentiated:
- the reference to the cleaned DOI was verified;
- no reference to the cleaned DOI could be found;
- the reference list and metadata of the given articles were not accessible, making it impossible to produce a statement about the correctness of the DOI.
98% of the cleaned cited DOIs proved as correct after the manual check. However, a closer investigation of the two DOI names which resulted incorrect showed that the reason is not to be imputed to the algorithm adopted. The DOI "10.1007/978-3-319-90698-0_26" should appear among the references of the article corresponding to the DOI "10.17660/actahortic.2020.1288.20", which unfortunately is behind a paywall and therefore inaccessible. This article was not the only one published on a toll-access journal among the 100 checked DOIs, but the University of Bologna, which the authors of this research are affiliated to, is not subscribed to the magazine in question, namely “ISHS Acta Horticulturae”. On the other hand, the DOI "10.1007/s10479-011-0841-3" should be cited by "10.1101/539833", but, although reported by Crossref, it does not appear in the original article, not only in its correct version but also in the invalid one, that is "10.1007/s10479-011-0841-3." with a point after the DOI. This mismatch can be explained as a processing error made by the platform that published the article, that is bioRxiv, since the same invalid DOI appears 118 times in the dataset analyzed and not even one in the articles that mention it.
Data Management Plan
The tense used in the DMP is the future, since it is developed before any data have been produced. Thus, the answers of the various points should reflect this.
The answers to the various DMP points were converted to the future tense.
Title: Classes of errors in DOI names: output dataset
Section 3.1.3: you have to specify the vocabulary used, avoiding the sentence "Couldn't find it? Insert it manually"
The vocabulary used for metadata has been specified. Since the dataset is published on Zenodo, the metadata about it is provided by Zenodo itself using the Datacite Metadata Schema (https://schema.datacite.org/).
Section 3.1.8: since you specified (in section 3.1.7) that you will use naming convention, you have to specify which naming convention is adopted.
The naming convention adopted was specified, in this way: "The field titles in the CSV file will be indicated in snake case, with each space replaced by an underscore. The first letter of each field will be capitalized, as well as the acronym "DOI" will always appear in capital letters. In addition, the DOI names reported in the cells of the file will always be in lowercase. Finally, whether or not a value belongs to a class will be indicated with 1 or 0, having the Boolean value of true or false respectively".
Section 3.1.9: here you should explain how to associate version numbers to your dataset, and not which version is currently in use.
This section has also been checked and edited. We used Semantic Versioning 2.0.0 (https://semver.org/) for both datasets.
Section 4.4: the answer provided is "[Other]" but not additional comment has been added there to clarify what that "[Other]" means.
We added a comment to clarify what "Other" means, that is "The dataset will be available on Zenodo under a Creative Commons Zero v1.0 Universal license".
Classes of errors in DOI names: code
Section 2.1: in this context the term "data" should be interpreted as "software". Thus, the question refers to if you are going to reuse existing software for accomplishing your goal. Under this meaning, please be aware that also the Section 2.2 and Section 2.3 could be populated with some information. Please notice that this interpretation of "data" as "software" may affect also other points of the DMP.
All libraries used within our code have been fully referenced, specifying which use has been made of them. References were made to the specific versions used, published on the Software Heritage Archive.
Section 3.1.9: here you should explain how to associate version numbers to your dataset, and not which version is currently in use.
The answer to this question has been modified. Semantic Versioning 2.0.0 (https://semver.org/) was used for both dataset.
Section 3.1.15: I do not believe you use/share Python Compiled Files (.pyc) but rather simple Python files (.py), right?
Right. The data format was corrected from Python Compiled Files (.pyc) to Python files (.py).
Section 3.4.5: in case you have developed unit tests to check the correctness of the code, then you have a documented procedure for quality. However, I do not know if you did tests or not.
The answer is no, the test-driven development procedure has not been adopted. However, we think it would have been better to adopt it. Although TDD requires tests to be developed before code, we believe that it still has value to develop them afterwards, to have a solid foundation in case of extensions or code changes. For this reason, we developed unit tests, using the Python built-in "unittest" framework.
Protocol
"The data needed for this work is provided by COCI as a CSV": it is not provided by COCI (which is a collection), it is provided by myself.
In our protocol, we removed all the mentions of COCI as a provider of the dataset and we substituted them with the reference to the dataset you provided on Zenodo.
"The records are provided on public licence by": change in "license in" and put the bibliographic reference in a shape it is clear (e.g. by using italic or other appropriate format).
The text was corrected as suggested. Any in-text reference now follows the APA citation style, and we added the reference list as the last section of the protocol.
"40'228": write "40,228" (the comma in English notation is visual separator).
As suggested, we used a comma instead of an apostrophe as visual separator for groups of thousands.
"there are two main classes of errors: author errors, made by authors when creating the list of cited articles for their publication, and database mapping errors, related to a data-entry error": it would be good to acknowledge here the fact that we have, in general, "human errors", i.e. those introduced by authors, editors and publisher representatives when managing the references - as explained during the workshop event.
In the newer version of the protocol we described the class of human-made errors, in addition to the two author errors and database-mapping errors classes.
"we manually isolated from a subset of 100 DOIs recurrent strings": how did you selected such 100 DOIs?
Since we didn't store the subset of 100 DOIs from which we extracted recurrent patterns of errors and we were not able to reproduce it, we removed any mention to this subset of DOIs and all we stated was that we did a manual identification of a subset of DOI names having recurrent prefixes and suffixes errors.
"eventual unwanted characters": remove "eventual"
We removed the occurrence of the word "eventual" in section 3.3 of the newer version of the protocol.
Software
The README.md file was added to the Github repository. In particular, the chapter "Getting started" describes how to reproduce the experiment, which methods to apply and in what order. The chapter "Hardware configuration" reports relevant specifications of the PC on which the research was conducted. The chapter "Results" refers to the website on which the outcomes' visualizations were published. Finally, the chapter "References" refers to the resources mentioned or linked, such as the DMP, the protocol and the article.
Please note: Due to technical problems attributable to Zenodo, it was not possible to publish the latest release of the code and then provide the DOI of the latest version. The technical problem is widespread and documented in the following issue: https://github.com/zenodo/zenodo/issues/2181. We do not know when it will be resolved, therefore, the DOI that refers to all versions of the software and that, by default, redirects to the latest version was used as a reference. Anyway, we will monitor the situation and make sure that the latest release is uploaded correctly to Zenodo as soon as the problem is resolved.
Update: the releases upload to Zenodo is working again and the latest version of the code, that is the version 1.1.0, was published with the following DOI: 10.5281/zenodo.4946380.
Article
All the mentioned issues have been addressed, correcting former inconsistencies and mistakes. In addition, the article has been updated with new content, including descriptions of the added methods applied to improve the research.
Dear @open-sci/grasshoppers,
Please find here attached my comments of all your material. You have to address all of them and, once finalised, to close this issue with a comment containing your reply to each of the points I have highlighted. There is no specific deadline to complete this task, thus please take your time.
Please, be aware that some modifications in some document may affect also modifications in other documents. As a final note, please remember to keep your notebooks up-to-date.
After closing this issue, please remember to update your
material.mdfile by specifying the references to the new version of all your documents.As usual, for further doubts, do not hesitate to contact me in the Signal group or just comment this issue here.
General comments from the presentation
You showed the result of your research by using a subset of the input citations. Would it be possible to have a complete coverage of the results by considering all the citations, instead of just a part of it?
It would be great to compare the results you obtained if you use only the procedure (i.e. regular expressions) described in Xu et al. vs all those that you obtained by using your updated methodology, i.e. that one which includes more regex as you have presented.
You said that it is pretty impossible that the process stop to compute the various values. But what does it happen if someone by accident switch off your computer while running the computation? Do you have a way to restart it from the last data produced, avoiding to start it again from the beginning?
An invited expert identified a bug in the code for cleaning the DOIs. Is it possible that this bug affected the results presented? If so, it would be necessary to recompute the whole data, of course.
Did you check, in some way, if the cleaning of a DOI produced a valid DOI which is indeed the article that is indeed cited by the citing entity? In other words, is it possible that your cleaning process produce a citation between two entities which is not the one that was originally defined in the citing paper?
DMP
The tense used in the DMP is the future, since it is developed before any data have been produced. Thus, the answers of the various points should reflect this.
Title: Classes of errors in DOI names: output dataset
Section 3.1.3: you have to specify the vocabulary used, avoiding the sentence "Couldn't find it? Insert it manually"
Section 3.1.8: since you specified (in section 3.1.7) that you will use naming convention, you have to specify which naming convention is adopted.
Section 3.1.9: here you should explain how to associate version numbers to your dataset, and not which version is currently in use.
Section 4.4: the answer provided is "[Other]" but not additional comment has been added there to clarify what that "[Other]" means.
Classes of errors in DOI names: code
Section 2.1: in this context the term "data" should be interpreted as "software". Thus, the question refers to if you are going to reuse existing software for accomplishing your goal. Under this meaning, please be aware that also the Section 2.2 and Section 2.3 could be populated with some information. Please notice that this interpretation of "data" as "software" may affect also other points of the DMP.
Section 3.1.4: the DataCite Ontology mentioned is not the official DataCite specification, bur rather a community effort to provide an OWL representation of (some portions of) the DataCite schema. Zenodo is not using SPAR DataCite Ontology in any way. Please correct.
Section 3.1.9: here you should explain how to associate version numbers to your dataset, and not which version is currently in use.
Section 3.1.15: I do not believe you use/share Python Compiled Files (.pyc) but rather simple Python files (.py), right?
Section 3.4.5: in case you have developed unit tests to check the correctness of the code, then you have a documented procedure for quality. However, I do not know if you did tests or not.
Protocol
"The data needed for this work is provided by COCI as a CSV": it is not provided by COCI (which is a collection), it is provided by myself.
"The records are provided on public licence by": change in "license in" and put the bibliographic reference in a shape it is clear (e.g. by using italic or other appropriate format).
"40'228": write "40,228" (the comma in English notation is visual separator).
"there are two main classes of errors: author errors, made by authors when creating the list of cited articles for their publication, and database mapping errors, related to a data-entry error": it would be good to acknowledge here the fact that we have, in general, "human errors", i.e. those introduced by authors, editors and publisher representatives when managing the references - as explained during the workshop event.
"we manually isolated from a subset of 100 DOIs recurrent strings": how did you selected such 100 DOIs?
"eventual unwanted characters": remove "eventual"
Software
The README.md in the GitHub repository does not contain an appropriate introduction to the software and, in particular, how to and in which order to call the various scripts to run the process correctly. In addition, you should also specify with which configuration (i.e. computer, processor, RAM, HD, etc.) you have run the scripts to get your final output, since this is crucial to foster reproducibility. Finally, if the software is somehow related with other documents (the protocol, the article, website, etc.), please mention them here in the README.md.
Article
"OpenCitations Index Of Crossref Open DOI-To-DOI References": it should be "OpenCitations Index Of Crossref Open DOI-To-DOI Citations"
"As the data provided by publishers to Crossref is not double- checked, the references integrated in COCI can still contain errors, which would mean a drawback for the goal of open scholarly citations": the references in Crossref may contain errors, but in COCI these are excluded.
"COCI validates the DOIs using the DOI API and provides a CSV list with all the DOI names that prove as invalid": it is not entirely true. COCI does not provide the DOI names that are invalid, simply they are not added in COCI. I (Silvio) provided such names. Then, I agree with you it would be good to share systematically this invalid DOI names within the OpenCitations website, something that can be done in the future.
"Consequently, the taxonomy...": you should mention in which section all the things you are summarising here will be presented. E.g.: "In Section 'Methods and Material' we present ...".
"1.223.296 DOI-to-DOI citations": please use the English notation for numbers, since "." is used to separate the decimal part of a number from the floating part. Thus, that number (and others, if any) should be written as "1,223,296".
"are highlighted in red in the table below" (and similar): when referring to tables, listings, figures, etc., you should avoid below/abobe/etc. but rather explicitly point to the particular table. E.g.: "are highlighted in red in Table 5".
"Table 5 Regular expressions...": explicitly add in the caption the meaning of the two colours adopted.
"As mentioned in the Materials and Methods chapter": it is a section, not a chapter.