In the code below the metrics summary file is added to the .uns layer after reading in the metrics_summary.csv file and formatting it using the process_metrics_summary() and more specifically the read_percentage() function. However, this causes issues where values that are not percentages are also divided by 100, resulting in incorrect values.
Below is an updated version of the read_percentage() function that first checks if there is a % sign in the value before dividing by 100. This seemed to resolve the issue on my side.
def read_percentage(val):
try:
if '%' in str(val):
return float(val.strip('%')) / 100
else:
return val
except (AttributeError, ValueError):
return val
Test:
Metrics file import:
I imported a metrics_summary.csv file from a cellranger multi run. All values are still correct.
Current code:
See how the 596 in the first row is converted into 5.96. Similar thing happens for the 165 value being turned into 1.65. This happens after running the read_percentage() function.
Updated code:
With the extra check for the % sign the issues described in the previous screenshot seem to be resolved while not affecting the conversion of percentage values.
In the code below the metrics summary file is added to the
.unslayer after reading in themetrics_summary.csvfile and formatting it using theprocess_metrics_summary()and more specifically theread_percentage()function. However, this causes issues where values that are not percentages are also divided by 100, resulting in incorrect values.https://github.com/openpipelines-bio/openpipeline/blob/196a69d6454916de7c5cf9927a8d9e7c16ca5f66/src/convert/from_cellranger_multi_to_h5mu/script.py#L131-L141
Below is an updated version of the
read_percentage()function that first checks if there is a%sign in the value before dividing by 100. This seemed to resolve the issue on my side.Test:
Metrics file import: I imported a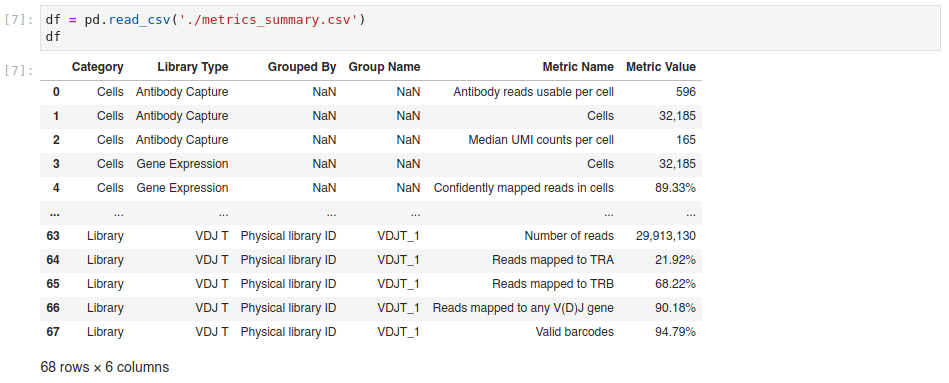 Current code:
See how the 596 in the first row is converted into 5.96. Similar thing happens for the 165 value being turned into 1.65. This happens after running the
Current code:
See how the 596 in the first row is converted into 5.96. Similar thing happens for the 165 value being turned into 1.65. This happens after running the  Updated code:
With the extra check for the
Updated code:
With the extra check for the 
metrics_summary.csvfile from a cellranger multi run. All values are still correct.read_percentage()function.%sign the issues described in the previous screenshot seem to be resolved while not affecting the conversion of percentage values.