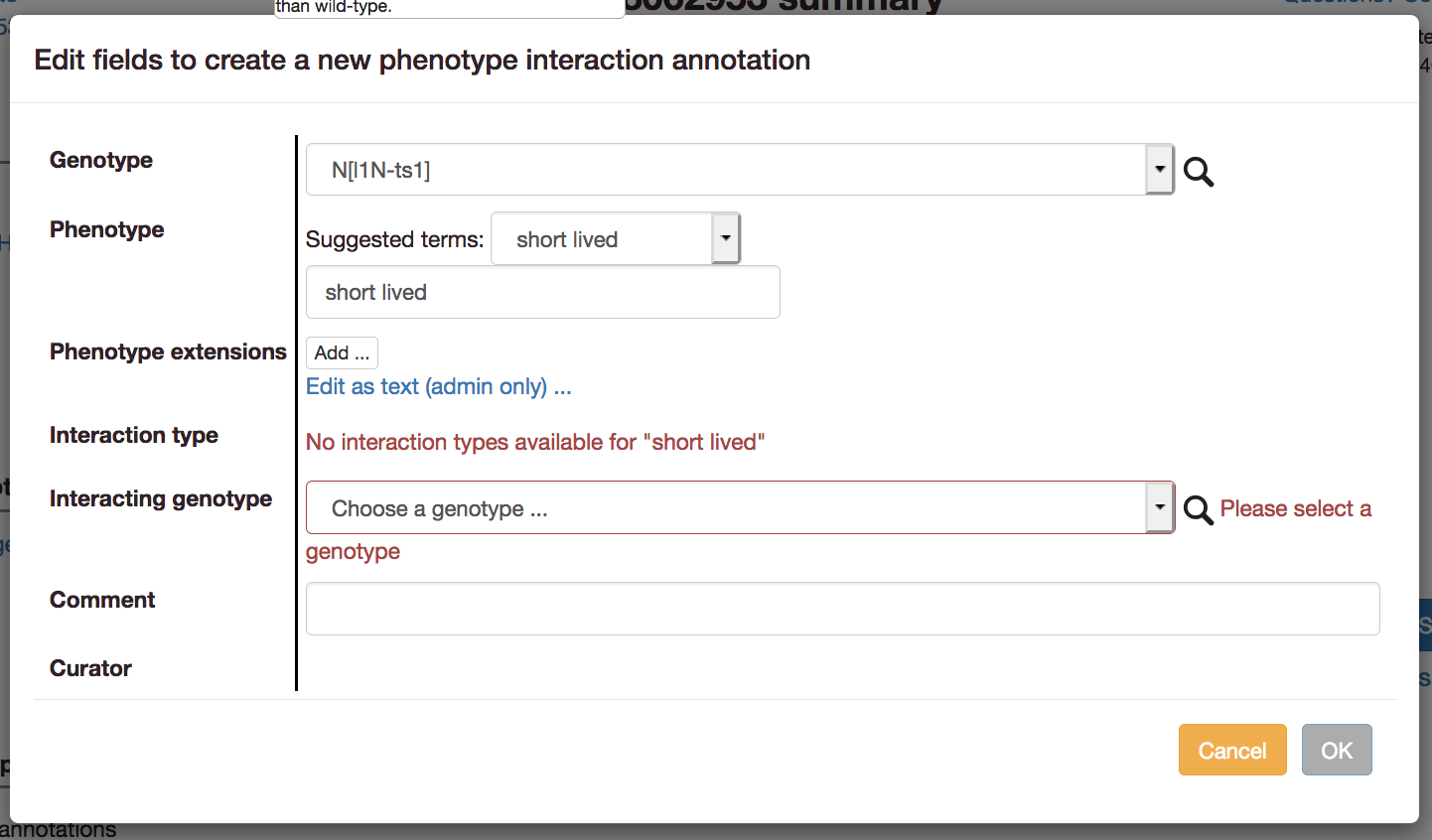
My memory isn't necessarily more trustworthy, but I think this option:
restrict possible phenotype given the selected interaction type
fits the envisioned workflow (at least as I understand it) a bit better. I'm going on the notion that it's
- pick genotype A 1a. (?) pick the affected phenotype of genotype A (optional? did we even actually decide on this?)
- pick an interaction type
- pick genotype B
- pick a phenotype for the genotype A / genotype B combination (is this step to be optional? maybe FlyBase needs it to be optional??)
If the above is generally accurate, then it would make sense to restrict the possible phenotypes at step 4 to ones relevant to the interaction type.
If we also include step 1A, then that phenotype selection/annotation should offer any phenotypes already annotated for genotype A, but also offer the option to add a new one (no restrictions ??). This could well be one of the interface complications that bogged it all down :P





(Following on from #1915)
Sorry I didn't take good notes on Skype. Please comment if I've got this wrong.
When editing interactions:
and either: