Thanks for the issue @Hoeze . Multi-dimensional bool indexing is definitely something we'd like to add.
How does your code differ from the proposals in https://github.com/pydata/xarray/issues/1887? In a brief look through the code — thanks for supplying it — I couldn't immediately see why we need a new dimension?
Currently, the docs state that boolean indexing is only possible with 1-dimensional arrays: http://xarray.pydata.org/en/stable/indexing.html
However, I often have the case where I'd like to convert a subset of an xarray to a dataframe. Usually, I would call e.g.:
However, this approach is incredibly slow and memory-demanding, since it creates a MultiIndex of every possible coordinate in the array.
Describe the solution you'd like A better approach would be to directly allow index selection with the boolean array:
This way, it is possible to 1) Identify the resulting coordinates with
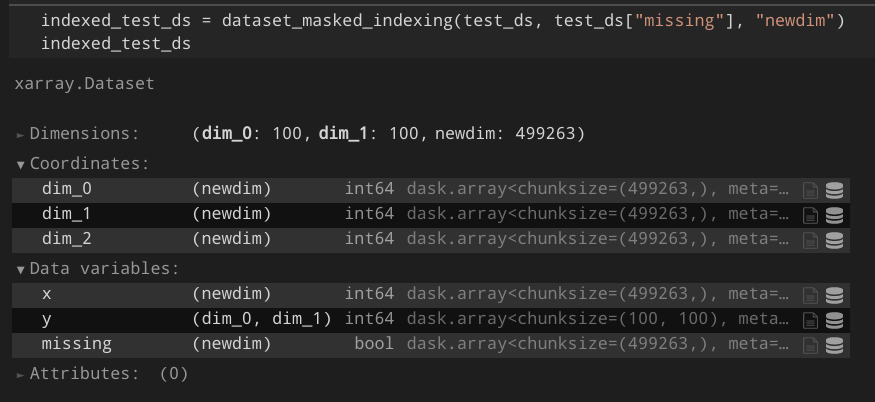
np.argwhere()2) Directly use the underlying array for fancy indexing:variable.data[mask]Additional context I created a proof-of-concept that works for my projects: https://gist.github.com/Hoeze/c746ea1e5fef40d99997f765c48d3c0d Some important lines are those:
As a result, I would expect something like this: