It looks like you have missed the step to pack the bytes of the overlay image into a bit array. There is a function in the numpy handler that does this - I think you can use it directly, or at least check how it can be done.
Closed gladomat closed 4 years ago
It looks like you have missed the step to pack the bytes of the overlay image into a bit array. There is a function in the numpy handler that does this - I think you can use it directly, or at least check how it can be done.
Good catch, thanks! Here are the additional lines of code necessary to make it work:
Import the pack_bits function:
from pydicom.pixel_data_handlers.numpy_handler import pack_bits
Reshape to 1D array and replace the to_bytes() call with this one:
im_data_over_bytes = pack_bits(im_data_over)
The result looks like it should now:

Hey @gladomat , I am still not able to overlay. The code ran fine without any errors, but the generated new DICOM is exactly similar to the original one. I couldn't see segmentation maps overlaid on the original DICOM. Any idea what might be happening?
Maybe your dicom viewer doesn't support overlays. The problem with overlays in dicom is that many software vendors don't even support it.
Thank you so much for you reply @gladomat . I am using Raidant, MicroDicom and Mango viewer but not able to see the overlay in any one of them. Do you have any suggestion or recommendation which DICOM viewer would be good to see the overlays?
Don't know about these, but I'm quite sure that Weasis shows overlays.
Thanks a lot @mrbean-bremen . At first I got this error _"UserWarning: Invalid value for VR CS: 'mask'. Please see https://dicom.nema.org/medical/dicom/current/output/html/part05.html#table_6.2-1 for allowed values for each VR.
warnings.warn(msg)"_
 I fixed this error by replacing "mask" with "MASK", all caps. Then the code worked fine.
I fixed this error by replacing "mask" with "MASK", all caps. Then the code worked fine.
Then, I tried troubleshooting the issue, and I found that the binarized segmented image has only 1's. Not a single 0 is present over there. Then I found that im_data_over = over.pixel_array has values of 4203 only. I am not getting what the issue is because the segmented DICOM image looks okay. I am attaching the segmented image for reference.

About the warning - yes we added checks for correct values, so that makes sense. Can't say anything about your actual problem without the dataset, though.
Thank you so much @mrbean-bremen. I am attaching the original DICOM and the segmentation map DICOM as well for your perusal.
Google Drive Link: https://drive.google.com/drive/folders/1u3i7uPObaSGLuRqx8-rfvH5atIMuX5tm?usp=sharing
I just had a look at your data (seem to have missed your post), and the problem is that your segmented image is not really mask data, but has different grey values - including the background, which is that 4203 you were seeing. And no, not all voxels have that value, just most of them (the background). Also, I'm not sure how that segementation relates to the original image. It has different width and height, and it is unclear where in the original image this would go.
The code above works for a binary segmentation mask that has the same size as the original image - not what you have got.
I'm trying to add an overlay to an already exhisting DICOM file. The idea is to have an original image, a segmantation of the original, and to add an overlay to the original image from the segmentation. Both images have the same dimension. I can't seem to be able to do it. The result is a garbled repeating pattern. I don't quite understand what I'm doing, so this is just an attempt. Any help is appreciated.
Here's a little bit of code I tried.
The result looks like this: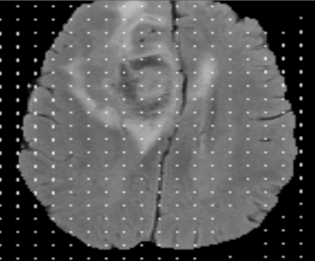
Edit: removed DICOMs. Sorry, they're not mine.