I have not checked, but from the looks of it, it looks like this is more an issue of matplotlib colormap usage. The colormap seems to be a gradient from white to red or green, and behaves accordingly. To get the desired visual effect, the colormap should be a gradient from transparent to red or green.
This so answer seems to achieve this effect by using RGBA colors and having a linspace as alpha along the colormap, from 0 at white (thus transparent) to 1 at green.
I think that using an alpha value will override the inherent alpha of the colormap, so you may need to set fill_alpha to None or "none", in that case you may need to clip the max of the linspace to what used to be fill_alpha.



 The code is here:
The code is here: 





Description of your problem
The
plot_gp_distfunction handlesfill_alphain a weird way (at least to me):Because the region between 1% and 99% and the region between 2% and 98% overlap, the
fill_alphaparameter might not have the intended effect. The stacking of multiple transparent regions will result in the center percentiles not having transparency at all? There are 40 semi-transparent rectangles stacked in the center. The stacking of colors and color mixing might also distort the colormap towards the center.Test-Graphs using rtcovidlive/rtlive-global with dataset from here, but heavily cropped.
What it looks like now (
fill_alpha=0.6):When setting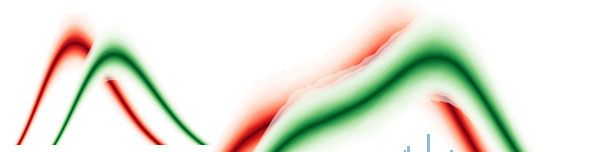
fill_alpha=0.6, fill_kwargs={"edgecolor":"none"}the hard lines disappear or get softer. Still not 60% transparent:What i expected (modified code with
fill_alpha=0.6, fill_kwargs={"edgecolor":"none"}):Code replacing these lines:
Fixing or changing this behavior has to be considered a BC-break, I guess. But maybe I'm also missing something and this style of "progressive transparency" was intended?
Versions and main components
conda env create -fenvironment.yml