Awesome! Nice job.
which you can find in the linked pull request.
Sorry, I don't see a link anywhere.
I don't use segments usually so forgive me if I'm misunderstanding, but would an alternative solution be to pre-allocate the final target array no matter what and fill it in with results? Then if a user wanted to provide the array that was being filled in they could provide an out keyword argument to resemble the standard practice in numpy?
Code Sample, a minimal, complete, and verifiable piece of code
Problem description
I am working on a software application that deals with large raster files (approximately 15000x15000 pixels). Recently, I encountered an issue where the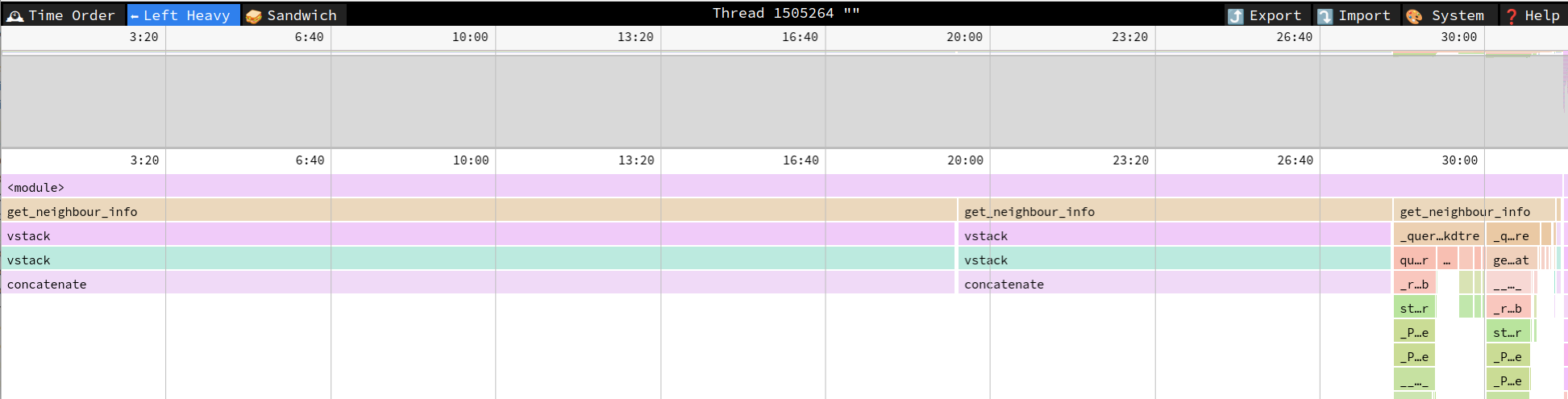
get_neighbour_infofunction was significantly slowing down, when I used multiple segments to fit the kd_tree lookups into RAM. The code snipped provided is a simplified benchmark demonstrating the issue. Upon profiling the code usingpy-spy, I discovered that the concatenation of segments was the cause of the slow down:As you can see, the current implementation results in many copies of the segments, which would even result in a peak RAM usage of 2 * total_size - segment_size.
Expected Output
To address this, a solution would involve pre-allocation of result arrays to avoid memcpys. Thanks to your habitable code base and excellent test coverage, I was able to quickly draft up a solution proposal, which you can find in the linked pull request. By pre-allocating the result arrays, most time is spent in the different workers querying the kd_tree, instead of copying data: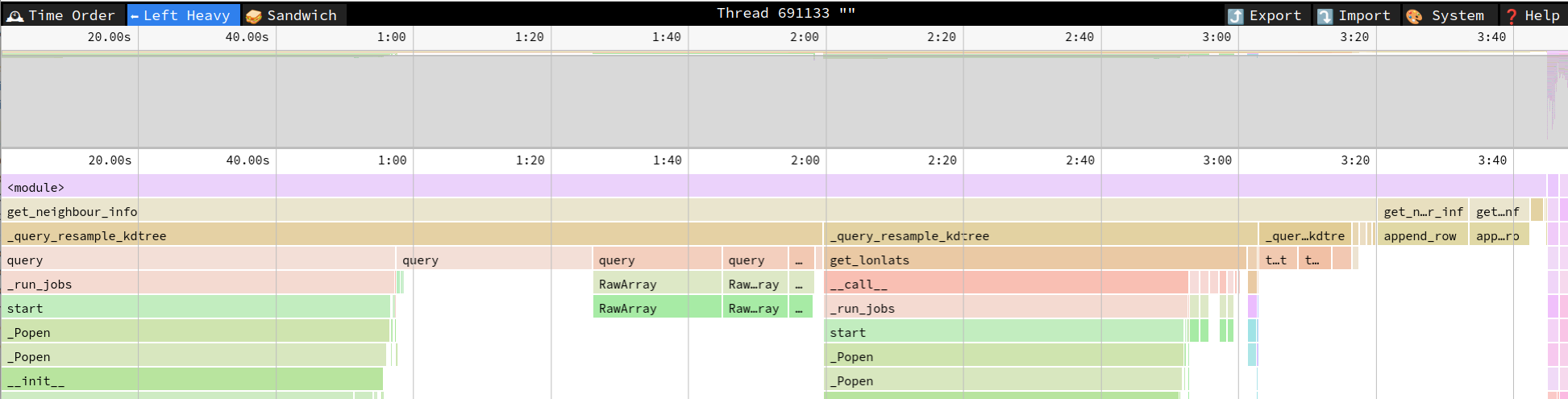
Also the execution time went down from ~35min to just ~4min.
Associated pull request
In this first draft I've opted for adding a simple pre-allocation size parameter to
get_neighbour_info, to avoid increasing the surface of the public API. However, another option would be to introduce a new method likeput_neighbour_infowhere the results are not returned, but provided as output parameters. This would give users even more flexibility such as specifying the result data type. However this comes at the cost of another public API function, and probably some significant refactoring ofget_neighbour_infoto avoid code duplication. What do you think?