Hey, I am also playing around this code. I have access to the eICU dataset. I have questions regarding the code and maybe u can help me out.
There are two files namely, "experiments.py" and "eICU_synthetic_dataset_generation.py". I am able to generate the data from the second one. I do not understand the use of "experiments.py" file for eICU datasets. There is no use of mmd or that in the "eICU_synthetic_dataset_generation.py" also.
Thank you.
I thought I'd share my experience with the MNIST data set and maybe ask some questions along the way. I used the csv version of MNIST. One of the first things I noticed was a problem with the labels. You might be able to train the model with the default settings, but in my case it did not process the labels correctly. So I found one way to fix this is to edit the mnistfull.txt settings file at experiments/settings/mnistfull.txt by changing the value of one_hot to true.
Next you copy your MNIST csv file to experiments/data/ directory. You also have to edit the code a little. Please note that since I did change the code in some of the files, the line numbers might not reflect the github archive. In any case, in line 16 of data_utils.py you want to comment out the imresize import because it is deprecated and not used for these settings. Like so:
#from scipy.misc import imresizeNext you have to fix some paths also in data_utils.py: Line 303:train = np.loadtxt(open('./experiments/data/mnist_train.csv', 'r'), delimiter=',')and line 308:np.save('./experiments/data/mnist_train.npy', train)Now something happens when you activate one_hot that detects the 10 labels corresponding to the 10 digits. However, experiments.py is set up to only read 6 or 3 labels. This will be the cond_dim settings variable. You don't have to set it in the settings file because it will automatically write it as 10 when you set one_hot to true. I'm not sure where it does this and it is worth further investigation. Suffice it to say you have to add some code to experiment.py at around line 103. You have to add another elif statement like so:
I was also having memory issues so I turned off mmd calculations according to this comment: https://github.com/ratschlab/RGAN/issues/16#issuecomment-359470046
Finally, you have to fix your print statements. The formatting expects a numerical value for mmd and that_np so look for a try/except block at around line 260 or so (in the archive it's at line 255). Before it you have to check to make sure that_np is initialized with the following code before the try:
Yes, that is you add another try before the try block with the print statements. Then you change the except print statement to the following:
print('%d\t%.2f\t%.4f\t%.4f\t%s\t%s\t %s\t %s' % (epoch, t, D_loss_curr, G_loss_curr, mmd2, that_np, pdf_sample, pdf_real))Basically, just making sure mmd2, that_np, along with pdf_sample and pdf_real are all formatted as strings.At this point, the model trained and I went for 500 epochs. Next, you want to generate some if not all of the digits. To do this, I did a couple of things first. In model.py, line 343, I change that line to this:
model_parameters = np.load(load_path, allow_pickle=True).item()You have to allow_pickle or the parameters stores as npy files will not load. Next, I wrote a script like so:You run this script just like you run experiment.py as such: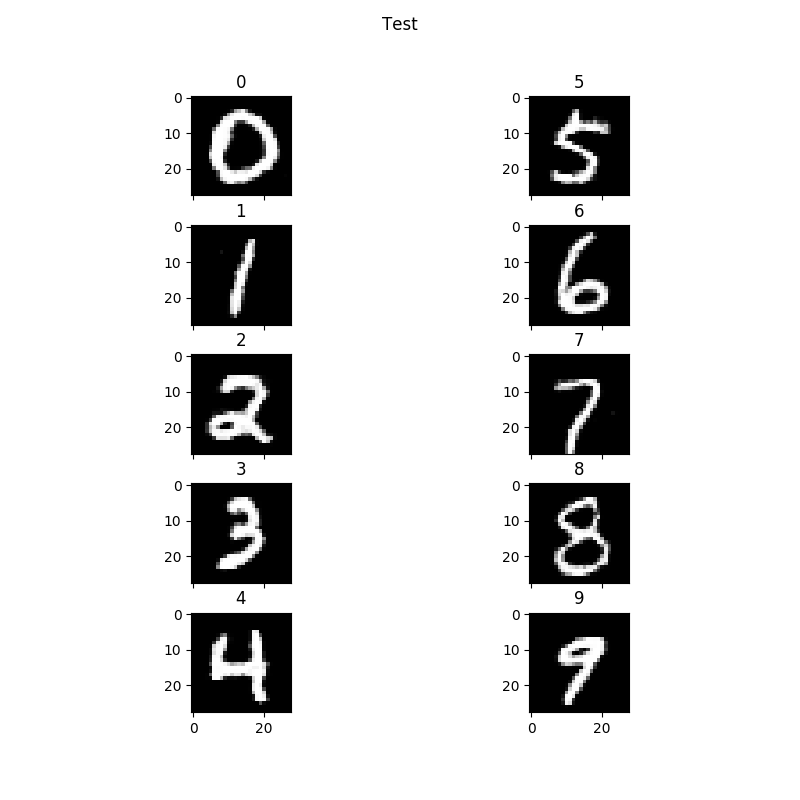
python script.py --settings_file mnistfullAlso, you probably don't need all the import statements. I just copy/pasted from experiment.py. Basically, your C_samples are the labels you input to the generator to tell it what digits to print. In this case, we are printing all ten digits. However they are formatted as a 10x10 numpy array each row being a digit with each column index indicating which digit it is by setting it to one. So for example if you want to print zero as the first digit, you set row zero column zero to one. If you want to print the third digit as 5 you set row three column five to one. In this case, I set every digit in order so 0,0 is 0, 1,1 is 1 and so on. Here is an example of my output:Now the minimum number of digits you can print is four based on how the plotting.py script is written. But you can print out any digit. Say you wanted to print out the digits 5309. You would use the same script as above but change the num_samples variable to 4. Create the csamples array with a shape of (4,10) and construct the array as:
Your output might look like this: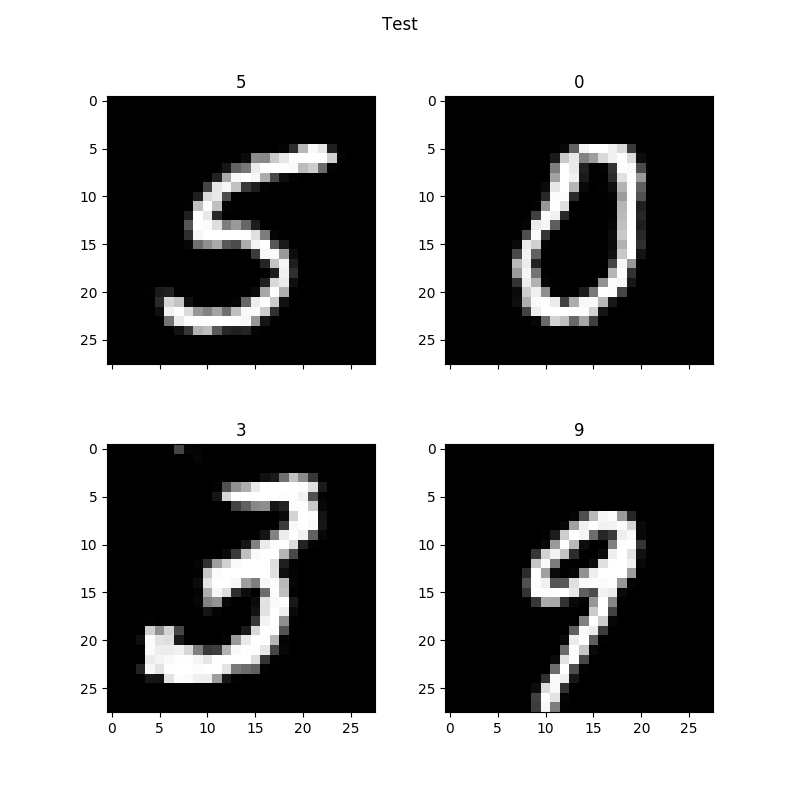
Note this is after 450 epochs. Well this was my experience with MNIST and this algorithm. Hopefully this will help someone along the way. I am also not clear from a theory perspective what the one_hot value does. Hopefully, I am not invalidating the results by the changes I made to the code. Thank you.