@RussellGrayxd I have just transferred your issue to this repo which is MODIStools repo. Cc @khufkens
Closed RussellGrayxd closed 4 years ago
@RussellGrayxd I have just transferred your issue to this repo which is MODIStools repo. Cc @khufkens
Before you use the package please inform yourself about the nature of the data product you use. The data is not wrong as you suggest, the interpretation of it is.
A tutorial is provided here:
https://daac.ornl.gov/resources/tutorials/MODIS-VIIRS_Webinar_20180816.html
The data is processed from raw sinusoidal MODIS tiles, as displayed below.
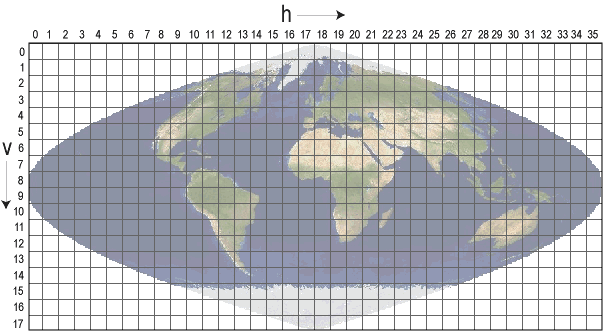
As you can see, on this projection the earth surface gets warped. And what you think is utter garbage is not, if you look in more detail to your data.
To limit bandwidth I use a MODIS land cover example, expanding the query to the maximum extent (100 km on a side) to illustrate my case (the same should apply to VIIRS data as it uses the same projection).
LC <- mt_subset(product = "MCD12Q1",
band = "LC_Type1",
lon = 121.1887,
lat = 23.8963,
km_lr = 100,
km_ab = 100,
start = "2004-01-01",
internal = TRUE,
progress = TRUE)
LC_map <- mt_to_raster(df = LC)
mapview(LC_map[[1]])This renders the following map, which nicely follows the Taiwan outlines. Note that the image is NOT square or rectangular and slanted due to the nature of the sinusoidal projection.

Sorry to bother you about this again. I used the code you added in the last email and it didn't re-project as yours did on my mapview. It's still the same as the first time I messaged about the issue. What am I doing wrong? I've read through the links you have sent me, and I understand now that the files are imported as sinusoidal tiles, but they don't say how to reproject them to normal format in R. Any help is appreciated. -RG On Saturday, December 14, 2019, 02:52:37 a.m. GMT+7, Koen Hufkens notifications@github.com wrote:
Closed #18.
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub, or unsubscribe.
Use:
LC_map <- mt_to_raster(df = LC, reproject = TRUE )
I was using a test setup in which this is the default, this is not the case for the normal package (due to overhead issues).
Thanks! On Saturday, January 4, 2020, 08:26:40 p.m. GMT+7, Koen Hufkens notifications@github.com wrote:
Use:
LC_map <- mt_to_raster(df = LC, reproject = TRUE )
I was using a test setup in which this is the default, this is not the case for the normal package (due to overhead issues).
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub, or unsubscribe.
Hi again, Can't seem to get the "reproject=TRUE" argument to work.
It just says: "Error in MODISTools::mt_bbox(xllcorner = df$xllcorner[1], yllcorner = df$yllcorner[1], : unused argument (transform = FALSE)"
I assume this is an error in the function itself.
| Also, is there any way to download the package with the function included in it? I've tried with remotes, devtools, and the basic "install.packages" and none have included it. Thanks --Russell J. Gray Wildlife Ecology & Conservation Consultant | 0934581341 | rgrayherpetology@gmail.com |
|---|---|---|
https://www.rjgrayecology.com/ | | 705 alligator ranch rd. New Smyrna Beach, FL, USA. 32168 |
On Sunday, January 5, 2020, 01:55:40 p.m. GMT+7, Russell Gray <russell_graypt@yahoo.com> wrote: Thanks! On Saturday, January 4, 2020, 08:26:40 p.m. GMT+7, Koen Hufkens notifications@github.com wrote:
Use:
LC_map <- mt_to_raster(df = LC, reproject = TRUE )
I was using a test setup in which this is the default, this is not the case for the normal package (due to overhead issues).
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub, or unsubscribe.
Please provide a reproducible example for me to work from.
This works (as per above):
devtools::install_github("khufkens/MODISTools")
library("MODISTools")
library("mapview")
LC <- mt_subset(product = "MCD12Q1",
band = "LC_Type1",
lon = 121.1887,
lat = 23.8963,
km_lr = 100,
km_ab = 100,
start = "2004-01-01",
internal = TRUE,
progress = TRUE)
LC_map <- mt_to_raster(df = LC, reproject = TRUE)
mapview(LC_map[[1]])| Thanks, I restarted R and deleted MODISTools/reinstalled and this time around it worked. --Russell J. Gray Wildlife Ecology & Conservation Consultant | 0934581341 | rgrayherpetology@gmail.com |
|---|---|---|
https://www.rjgrayecology.com/ | | 705 alligator ranch rd. New Smyrna Beach, FL, USA. 32168 |
On Thursday, February 6, 2020, 03:15:01 p.m. GMT+7, Koen Hufkens <notifications@github.com> wrote: This works (as per above): devtools::install_github("khufkens/MODISTools") library("MODISTools") library("mapview")
LC <- mt_subset(product = "MCD12Q1", band = "LC_Type1", lon = 121.1887, lat = 23.8963, km_lr = 100, km_ab = 100, start = "2004-01-01", internal = TRUE, progress = TRUE)
LC_map <- mt_to_raster(df = LC, reproject = TRUE) mapview(LC_map[[1]])
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub, or unsubscribe.
I've queried several NDVI and landcover products at all bands over the past few days using the central lat/lon of Taiwan. Every single one of the products is incorrect. They don't represent the outline of the country, and they make no logical sense. I'm not sure where the landsat images are coming from, but it isn't Taiwan.
Here is a working example: library(MODIStools) library(mapview)
NDVI <- mt_subset(product = "VNP13A1", band = "500_m_16_days_NDVI", lon = 121.1887, lat = 23.8963, km_lr = 20, km_ab = 100, start = "2019-01-01", internal = TRUE, progress = TRUE)
NDVI_r <- mt_to_raster(df = NDVI)
mapview(NDVI_r[[1]])