Following up on this, is there anything I can do to help, like suggest additional reviewers?
Closed goodmansasha closed 6 years ago
Following up on this, is there anything I can do to help, like suggest additional reviewers?
Thanks for your submission @predict-r! And sorry for the slow reply, this fell through the cracks for a week. This is a good fit and I'll go ahead and find reviewers.
Here are the results of our automated tests via goodpractice::gp(). You have good code coverage (100% not required, but this helps rviewers see where they might look for edge cases that need coverage.) Most of the other stuff should be straightforward and I'll go ahead and look for reviewers. You can make those changes either before or after you get your first-round reviews.
I suspect you'll have trouble getting a >50MB .jar file onto CRAN. I'll leave this until we have a review from Java-experienced reviewer, but I suspect you may have to pursue an approach such as packaging the .jar file in a separate package and listing it as a dependency (see CRAN polices), file or having a function like install_tika() in the package that downloads the file before the package is functional.
── GP rtika ─────────────────────────────────────
It is good practice to
✖ write unit tests for all functions, and all package code in general. 92% of code lines are covered by test cases.
R/tika_html.R:12:NA
R/tika_json.R:17:NA
R/tika_text.R:13:NA
R/tika_xml.R:13:NA
R/tika.R:128:NA
... and 7 more lines
✖ use '<-' for assignment instead of '='. '<-' is the standard, and R users and developers are used it and it is easier to read your code for them if you use '<-'.
inst/doc/text_processing.R:121:11
✖ avoid long code lines, it is bad for readability. Also, many people prefer editor windows that are about 80 characters wide. Try make your lines shorter than 80 characters
inst/doc/text_processing.R:12:1
inst/doc/text_processing.R:58:1
inst/doc/text_processing.R:62:1
inst/doc/text_processing.R:84:1
inst/doc/text_processing.R:106:1
... and 56 more lines
✖ omit trailing semicolons from code lines. They are not needed and most R coding standards forbid them
inst/doc/text_processing.R:36:66
✖ avoid sapply(), it is not type safe. It might return a vector, or a list, depending on the input data. Consider using vapply() instead.
R/tika.R:185:18
✖ not use exportPattern in NAMESPACE. It can lead to exporting functions unintendedly. Instead, export functions that constitute the external API of your package.
✖ fix this R CMD check NOTE: installed size is 53.3Mb sub-directories of 1Mb or more: java 53.1Mb
─────────────────────────────────────────────Reviewers: @juliasilge @davidgohel Due date: 2018-03-05
Thank you @juliasilge and @davidgohel for agreeing to review! Reviews will be due 2018-03-05.
@predict-r You can add the review badge to your README now if you wish. It will auto-update with review status:
[](https://github.com/ropensci/onboarding/issues/191)Thanks! I'll go ahead and add the badge then. I'll patch some of the goodpractice issues listed above.
Thanks for pointing out the CRAN situation. I can create another repository with the Jar, and call it 'tikajar' . That package won't update often, so maybe CRAN will accept it. If not, the package can be left on github and downloaded with devtools.
Okay, I removed the .jar file. It is in a new package on github called 'tikajar' (see: https://github.com/predict-r/tikajar ). Since its a key part of 'rtika', I added the review badge to tikajar as well. Let me know if that's an issue.
Hi, here is my review:
Please check off boxes as applicable, and elaborate in comments below. Your review is not limited to these topics, as described in the reviewer guide
The package includes all the following forms of documentation:
URL, BugReports and Maintainer (which may be autogenerated via Authors@R).davidgohel: I tested with 1204 documents (my admin folder). I got 20.354 sec. with 8 threads and 36.376 with 1 threads. This is really good (it tooks 20 second to get my whole admin directory scanned and returned as an R object).
I was expecting a faster execution with 8 threads (not 8 times faster but at
least twice minimum). I used the profiling tools of RStudio and could see that the
function spent 1/3 of time executing .rtika_readFile() and 1/3 executing enc2utf8().
I think that starting JVM with arg -Dfile.encoding=UTF-8 instead of using enc2utf8
could be a solution (not tested here, but was the solution in past experiences...).
davidgohel:
There is no Code of conduct (but it is only recommanded in the guidelines)
Authorship: in field Author I recommand you the format with Authors@R: c(person(...).
One benefit for example is you will ensure programmatically that the roles are correct.
Package dependencies: in R/tika.R (l179), you are using utils but it is not in Suggests.
Package stats is used in R/tika.R (l130) and should be declared in Imports.
The way you are using Suggests and requireNamespace instead of Imports
is unconventional to me but I think it is valid.
SystemRequirements: I don't think it will be accepted as is. (2 months ago, R Core Team
standardized java value), it should be java (>= 7). Also CRAN is asking to double check the
version of java - something like:
.onLoad= function(libname, pkgname){
.jpackage( pkgname, lib.loc = libname )
jv <- .jcall("java/lang/System", "S", "getProperty", "java.runtime.version")
reg <- "([0-9]+)\\.([0-9]+)\\.(.*)"
majorv <- as.integer(gsub(reg, "\\1", jv))
minorv <- as.integer(gsub(reg, "\\2", jv))
if(majorv == 1 ){
if( minorv < 7 ) {
stop("java version should be at minimum version 7 (>=1.7)")
}
}
}* checking examples ... OK
Examples with CPU or elapsed time > 5s
user system elapsed
tika 11.477 0.636 8.347
tika_text 5.786 0.328 3.875
tika_json 5.582 0.313 4.163
tika_xml 5.387 0.299 2.787
tika_html 5.224 0.308 2.812As the unit tests are there and cover almost
100%, I suggest you use \donttest{} for the 5 examples.
R CMD check did not succeed (via RStudio ckeck button).
I am running R 3.4.3 on OSX High Sierra 10.13.3.
Some tests are failling:OK: 49 SKIPPED: 0 FAILED: 5
1. Error: tika parses multiple local files (@test_tika.R#58)
2. Error: tika outputs NA with a path to nowhere in right order (@test_tika.R#86)
3. Error: tika outputs parsable xml (@test_tika.R#119)
4. Error: tika outputs parsable html (@test_tika.R#135)
5. Error: tika outputs parsable json (@test_tika.R#152) The fun fact is that these are sucessful when launched via RStudio test button... Maybe that's because of my environment. You can use that bit of code to check the package with R for OSX on travis:
https://github.com/davidgohel/gdtools/blob/master/.travis.yml#L5
Estimated hours spent reviewing: 4
Congratulations for that package, it brings someting new and really useful in the R data importation ecosystem.
It works well and is simple to use. I appreciated the help files. Vignettes are giving lot of examples and shows how to use the results.
A minor modification might be necessary: starting JVM with option -Djava.awt.headless=true,
this should ensure it will run on a server with no graphical environment.
More a wish than a comment... I would have used rJava API in order to avoid system calls.
I believe this could cause troubles in corporate environments where IT teams are sometimes
using tools that deny access to command line for some users.
I don't know tika but the rJava call should be something like:
.jcall("org.apache.tika.wrongbatchclassname","V","main",.jarray(list(...), "java/lang/String")About the size of the jar file, I did not read anything that says it's an issue (it will be if often updated) but you could ask to CRAN-submissions@R-project.org or r-package-devel@r-project.org what is the recommanded strategy.
Hope you will find that review useful :)
David
Thank you David for the very helpful comments. I've attempted to address your concerns in the 0.1.5 version, and hope they meet your approval. I'm happy that the batch speed was good, and have sped up the package a bit more this time.
I got 20.354 sec. with 8 threads and 36.376 with 1 threads.
Yes, I wish there was a better improvement in speed with that many threads. It seems like the speed per thread shows sharply diminishing returns, unfortunately (perhaps this is because of file system read speed maxing out, but that is just a guess). Your experience had led me to increase the default number of threads to two. Tika's default is the number of processors minus one, according to the batch processor config file. If you think I should use that instead, I will.
I used the profiling tools of RStudio and could see that the function spent 1/3 of time executing .rtika_readFile() and 1/3 executing enc2utf8().
This was very helpful. Your comment led me to read the Tika source code more carefully. I am happy to report that Tika outputs UTF-8 now. It was not immediately apparent, because the original Tika used UTF-16. However, the newer batch processor's FSOutputStreamFactory defaults to UTF-8. There is no need to create a special config file or anything, so I removed the offending enc2utf8(). This will speed up the package quite a bit! Thanks.
There is no Code of conduct
Thank you for reminding me. I've added one.
Authorship formatting
I've updated the authorship to use 'person' functions, as you suggested.
Package dependencies: in R/tika.R (l179), you are using utils but it is not in Suggests. Package stats is used in R/tika.R (l130) and should be declared in Imports.
The utils and stats packages are now declared in Imports.
Java SystemRequirements
I've updated the Java declaration to use the standard CRAN version style you mentioned. I've also included an .onLoad() function to verify that java is available and is the appropriate version.
The run examples are too long to be executed
These have been stopped using \donttest{} statements. I also made a change to the tests to speed them up, that you may or may not approve of. Instead of downloading files for the test each time, which took fifteen seconds or so, I moved a few different files into the test directory and now test on those.
R CMD check did not succeed
After several hours, this has been fixed on both linux and OS X. On my OS X laptop, I was able to replicate the error by using the system2 command to invoke java. The problem was that system2 stops Tika from finishing in some cases, terminating the process in the middle. The problem was intermittent. It did affect the same tests as you reported. By making the 'sys' package a requirement instead of suggestion, and using sys::exec_wait, the issue is fixed on my end. The .travis.yml is updated to test on both osx and linux now, and is passing. As a side effect, sys::exec_wait speeds up rtika by up to five seconds on my laptop.
A minor modification might be necessary: starting JVM with option -Djava.awt.headless=true, this should ensure it will run on a server with no graphical environment.
Thank you for this, and it has been implemented.
More a wish than a comment... I would have used rJava API
Thank you for letting me know. I agree that the rJava package could help with certain security issues. It would definitely bring speed benefits over repeated command line invocations. However, it would take time to to dive into the Tika codebase, implement it and test it. Perhaps my main issue is that I've had a some recurring frustration over the years installing rJava. While I'm not opposed to rJava, I would prefer not to add the extra complexity in this first version. If the rtika package is popular at all, I will add rJava.
About the size of the jar file, I did not read anything that says it's an issue (it will be if often updated) but you could ask to CRAN-submissions@R-project.org or r-package-devel@r-project.org what is the recommended strategy.
Thank you, I will look into the .jar size issue next!
I hope I've addressed your questions and comments, to your satisfaction.
Awesome, thank you.
There are 2 NOTE(s) left that could be suppressed:
https://travis-ci.org/predict-r/rtika/jobs/344591725#L349
rtika.pdf, R will built it from the man/ directory. packageStartupMessage() that should not be called from .onLoad(), using warning() instead should solve that.Thanks for your thorough review, @davidgohel!
David, I fixed the issue with the two NOTES.
Thank you Sasha
Hello! Here is my review. 🙂
Please check off boxes as applicable, and elaborate in comments below. Your review is not limited to these topics, as described in the reviewer guide
The package includes all the following forms of documentation:
URL, BugReports and Maintainer (which may be autogenerated via Authors@R).Estimated hours spent reviewing: 3
I have had HORRIBLE EXPERIENCES with installation and usage for Java-dependent R packages :scream::scream::scream: so I was full of dread I was asked to review this package. However, I am very happy to report that I was able to install and use this package without any trouble at all. Great job on this package, its test coverage, and usability!
library(rtika)
library(tidyverse)
library(tidytext)
my_path <- "~/Documents/Very Ancient Stuff/Teaching & Astronomy/"
batch <- paste0(my_path, list.files(path = my_path))
text <-
batch %>%
tika_text()
tidy_df <-
data_frame(file = batch,
text = text) %>%
unnest_tokens(word, text) %>%
count(file, word, sort = TRUE) %>%
anti_join(get_stopwords())## Joining, by = "word"tidy_df## # A tibble: 3,652 x 3
## file word n
## <chr> <chr> <int>
## 1 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/phy… students 116
## 2 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/phy… writing 68
## 3 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/phy… physics 60
## 4 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/Uni… universe 57
## 5 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/Uni… galaxies 45
## 6 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/Spe… science 43
## 7 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/phy… assignm… 40
## 8 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/phy… class 40
## 9 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/Uni… http 36
## 10 ~/Documents/Very Ancient Stuff/Teaching & Astronomy/Spe… god 33
## # ... with 3,642 more rowsmetadata <-
data_frame(raw = batch %>%
tika_json()) %>%
filter(!is.na(raw)) %>%
mutate(JSON = map(raw, jsonlite::fromJSON, flatten = TRUE)) %>%
pull(JSON) %>%
map_df(function(x) { x[1:4] %>% head(1) })
metadata## Application-Name Author Company
## 1 Microsoft PowerPoint Robert Silge Robert Silge
## 2 Microsoft Excel Robert Silge
## 3 Microsoft PowerPoint Robert Silge UT Southwestern
## 4 <NA> tech <NA>
## 5 Microsoft PowerPoint Information Technology Calvin College
## 6 Microsoft PowerPoint dhaarsma Calvin College
## Content-Length Content-Type Creation-Date
## 1 5082624 <NA> <NA>
## 2 30720 <NA> <NA>
## 3 8350720 <NA> <NA>
## 4 160175 application/pdf 2001-07-20T19:21:49Z
## 5 1538048 <NA> <NA>
## 6 9430528 <NA> <NA>When I run goodpractice:gp() on the current development version, I get this note about the new tikajar dependency:
fix this R CMD check NOTE: Namespace in Imports field not imported from: ‘tikajar’
All declared Imports should be used.I have not built a package that uses a .jar file in this way so I do not know how to address this TBH.
I have some suggestions around code style to place this package more squarely within R community norms as expressed, for example, in Google's R Style Guide or the tidyverse style guide. I don't know how annoying you will find this :flushed: but the value here is that other contributors will be able to read and understand your code more easily. Here are some big picture thoughts:
-> for assignmentThis is especially important in the README and vignette where the code is exposed to more people. I am about to submit a PR with changes to parts of the package but I wasn't able to go thoroughly through all the code today. This is worth it to make this a welcoming R community package.
The vignette currently focuses on text mining of the text that we can get out of this package. Almost the entire vignette focuses on text mining (using base R and some data.table) exploratory data analysis, which is not what this package is about; only the very first code chunk uses functions from rtika at all. The text mining community in R is active and healthy (I am part of it!) and there are lots of opportunities to learn about how to do text mining once you have text; I would suggest that a vignette for a package like rtika is not really the right spot for this kind of analysis. When I am learning how to use a new package, what I want from the vignette is how to use the functions in that package. I would love to see, instead, a more detailed exercise of how to use the actual rtika functions and what they can do. There is nothing in the vignette about handling metadata from multiple sources, for example. The current material that is in the vignette would be more appropriate for a blog post, or perhaps a secondary, supportive vignette.
I'd also like to address the tone of the vignette a little. I know it is so hard to write for audiences that you aren't quite sure where they are! :smiley: I am 100% in favor of writing accessibly without jargon so that people can get started, but I want to make sure this vignette doesn't come off as condescending. For example, if a user has managed to install the correct version of Java and is attempting to walk through a vignette, they probably know what regular expressions are.
Thanks for your thoughtful review, @juliasilge! I especially concur with your comments on the vignette.
Regarding the NOTE: I think the correct field for a package providing .jar files is Depends:. Perhaps @davidgohel can confirm.
That's because tikajar is not imported in NAMESPACE (but is in Imports in DESCRIPTION)
Adding #' @import tikajar somewhere in R/ should solve the problem.
@juliasilge Thank you for your helpful comments about making the package feel more welcoming. Based on the feedback here, we need a new vignette. It should demonstrate how to use the actual rtika functions and to show what they can do. It is also an opportunity to show the breadth of uses.
I'll merge your pull request, and go through each point in a coming response.
This morning, in the meantime, I'm brainstorming the vignette. If people know a good vignette to study, let me know please!
I'm thinking of a survey of uses and data. It will have more than five examples, each brief, each focused on a distinct case, each using a cool data set, and each following the 'tidy tools manifesto' principles of using pipes and common formats.
The more diverse the examples, the better. I'm thinking :
As for data, my data sources feel too similar to one another and am open to suggestions, especially anything from the Internet Archive (https://archive.org/ ). My initial ideas:
More diverse data examples feels more welcoming, especially documents appealing to the humanities and arts.
Thank you.
Around five example data sets would be plenty for a vignette, I think! Even three small case studies would be good. I would be thoughtful in what tooling you choose to use in the vignettes. For example, if you wanted to use the Stanford CoreNLP in the vignette, then that package would need to be in Suggests, and that package is a BEAR 👿 to get to install on Travis. I know this from experience. I'd try to stay quite lean and simple in what you use in example analysis in the vignettes and keep complex dependencies for demonstration blog posts.
Two examples packages that I think are good to look to for how to handle vignettes in this case are gutenbergr (downloads public domain texts from Project Gutenberg) and fulltext (searches and downloads full text for journal articles).
@juliasilge, I've implemented changes to address your helpful comments.
Issue with R CMD check
To avoid this issue, and make things simpler until CRAN complains, I've put the .jar file back into the 'rtika' package for now. So, the issues with importing 'tikajar' is gone on my end.
@davidgohel said: "About the size of the jar file, I did not read anything that says it's an issue (it will be if often updated) "
The size might not be an issue unless the package is updated frequently, and I hope to update it infrequently.
Code style
The 'styler' package came in handy. I've tried to make the README and vignette use a standard style throughout.
Thoughts on the vignette
I've written an introductory vignette, and removed the last one. This one focuses on how to use the package functions and process the metadata.
It is on the Github Pages site: https://predict-r.github.io/rtika/articles/rtika_introduction.html
I tried to keep it light and easy to read, and hope it meets your approval for a package vignette. The site was built with pkgdown::build_site().
By the way, I noticed that the review badge was not loading here, so had to remove it.
Thanks for your response, @predict-r! @juliasilge, let us know if these changes have addressed your comments and check that box off if so, let us know what else remains if not. I see @davidgohel already has checked off his box.
FYI, we had a temporary snafu on the service serving badges. It should be back now.
Thank you.
Ah, I love the new vignette!!! 💕 It is full of so much useful, detailed information. This is going to be a great resource for users. I also am happy to see the other issues in the review addressed; I reviewed the package once more, did R CMD check and goodpractice::gp() again, and I have checked off the box in my review.
The only thing significant remaining in the checks is the NOTE about the size of the package.
I do want this package to be a useful resource, and am so glad you liked the vignette!
The only thing significant remaining in the checks is the NOTE about the size of the package.
I will keep trying, but CRAN never got back to me at all.
Has anybody seen a package that downloads the required resources after package installation? There are Apache mirrors for Tika, such as http://mirrors.ocf.berkeley.edu/apache/tika/tika-app-1.17.jar .
Thanks all for great reviews and follow-up, @juliasilge @davidgohel and @predict-r. We can accept the package as-is from our perspective, the size/CRAN issue isn't a precondition on our front, but it makes sense to work out the approach first. I'll be happy to do a full editors' check if you make major changes on this front.
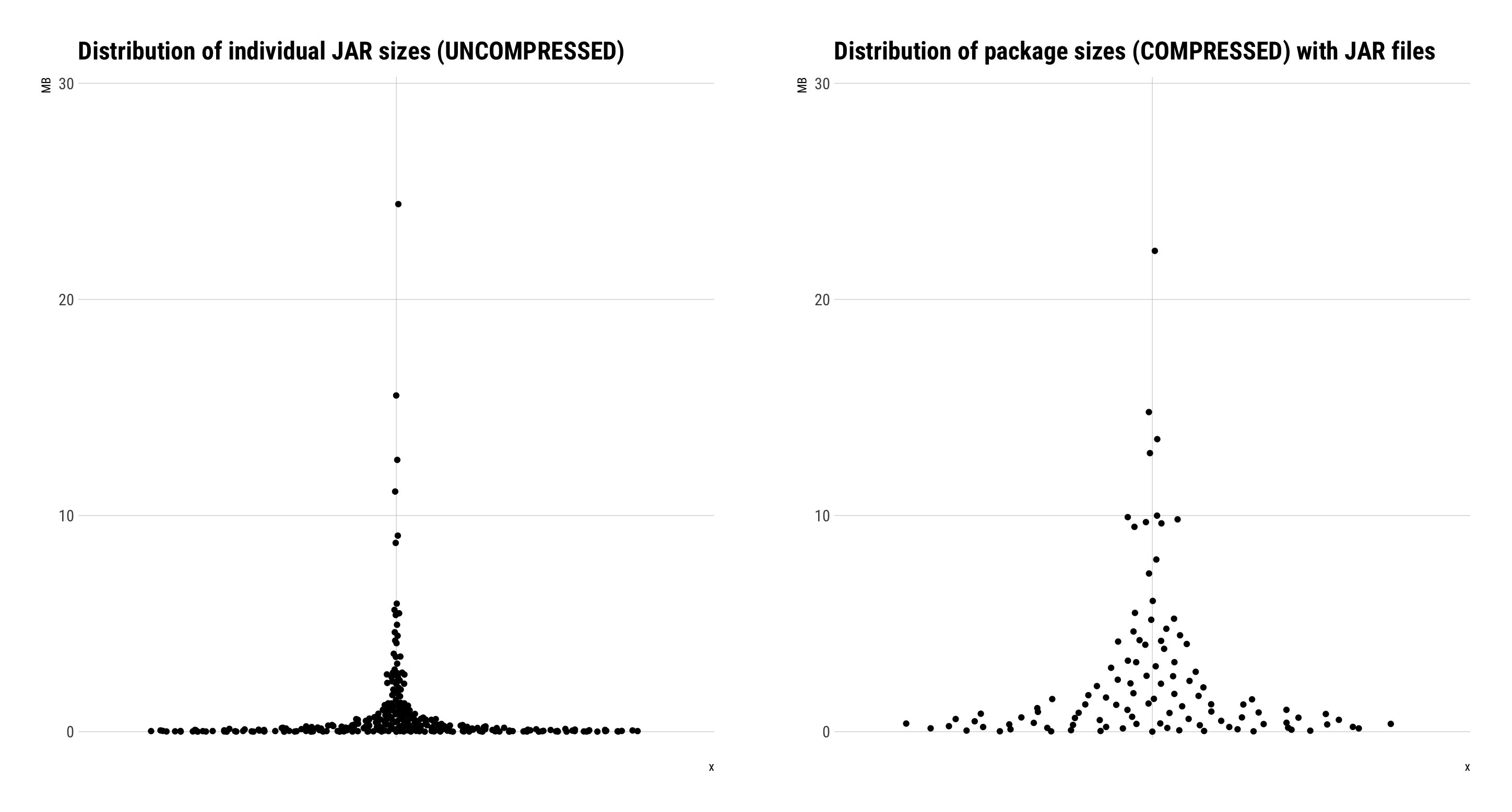
Courtesy of great friend of rOpenSci Bob Rudis, here's the distribution of .jar file sizes on CRAN. the Tika .jar would be larger than any of them, but isn't a different order of magnitude. So you could go ahead and submit tikajar to CRAN and see the what the response is, then submit rtika if it goes in.
If not, I believe having a package automatically install resources on installation is a violation of CRAN policies in most cases. The few exceptions I know of relate to license issues or platform-specific installs of compiled binaries. But plenty of packages have functions to install system dependencies that users run after install, such as keras::install_keras(). So having a tika::install_tika() function that downloads the .jar file (probably using rappdirs::user_data_dir() to select the install path) is a reasonable alternative strategy. In this case you will want to mark all examples as \dontrun{} and use skip_on_cran() for your tests so the package test properly on CRAN. You can still run install_tika() and run the tests on Travis.
We can accept the package as-is from our perspective, the size/CRAN issue isn't a precondition on our front
I've slept on it, and am ready to give up on CRAN for now. The response from Uwe Ligges at CRAN is:
In principle a CRAN package should not exceed 5MB, 50 seems to be too much.
Since the Tika .jar is over 55.7MB (49.5MB compressed), there is no way to distribute the .jar on CRAN without violating that principle.
So, once accepted this package would be on both github and ropensci! I'm happy with that.
@noamross Thank you for showing a good way, and offering to do a full editor's check. Uwe from CRAN had a similar suggestion:
I wonder if you cannot host that software externally and provide some infrastructure that allows to install the jar files later on if a user requests it from an external source?
The way you suggested, using rappdirs::user_data_dir() to get a writable directory, sounds fine. However, I still want to move forward and keep the package simple to install and uninstall.
Perhaps we can return to this issue and I could requests an editorial review in the future? Maybe there could be two versions of the package--one without the .jar?
I advise against two packages. It will be confusing for users and make more effort for maintenance. I think an install_tika() function makes for a fairly simple experience - you can print a package startup message if the .jar if missing, and a similar informative error for any Tika-calling function. Plus you can update the Tika version by one line of code - the download URL - in the package. CRAN publication will maximize your user base, as this package will of a great use to many people.
I'm OK with marking this as "accepted" now, but I would prefer that if the plan is to change the user experience in this way soon, we do so before we move forward with blog posts and such.
Alright, I’ll make the install script. :)
I've implemented install_tika().
However, along the way, I ran into a big Apache Tika bug! It was in the batch processor. The good news is that I reported the bug, and it was immediately patched! See [1].
The bug, combined with install_tika(), breaks rtika on OS X.
A potential issue is the next major release of Tika with the patch, Tika version 1.18, will be released soon, but I'm not sure exactly when...I've asked.
In the meantime, what about having install_tika() get the patched development version directly from Apache? See [2]. This would come with warnings that it is a development version.
If people have thoughts on installing a development version of Tika, I'd like to hear them.
[1] https://issues.apache.org/jira/browse/TIKA-2604?page=com.atlassian.jira.plugin.system.issuetabpanels%3Acomment-tabpanel&focusedCommentId=16393363#comment-16393363 [2] https://builds.apache.org/job/tika-branch-1x/10/org.apache.tika$tika-app/
P.S. I'm kinda proud of finding the bug!
👏 Good work finding that @predict-r! I had problems with another program crashing recently because of spaces in file paths, so I'm glad this bug is squashed.
We'll see what the response is from the developer, but based on this email thread it seems that 1.18 is only a week or two off. If it were months away I would suggest an jumping ahead. I would say it's fine to the patched version on GitHub now, (esp. given that user_data_dir() is likely to have spaces on many machines), but wait until 1.18 before CRAN submission. GitHub packages are generally perceived to be "development" versions. You can put a note about the patched version in the README.
I will take a careful look at the remaining changes to wrap this up later this weekend.
Thanks ! The install_tika() is temporarily set to download the development version. The package is passing Travis.
Approved! Great work, and thanks a lot @juliasilge and @davidgohel for your reviews! The install_tika() functions and docs are looking good, and I commend your good use of assertions and failsafes.
To-dos:
[x] Transfer the repo to the rOpenSci organization under "Settings" in your repo. I have invited you to a team that should allow you to do so. You'll be made admin once you do.
[x] Update badges, CI and coverage links, and URLs to point to the new repo address.
If you would like to, and if your reviewers agree, you may choose to credit their reviews in the DESCRIPTION file by adding them as 'rev'-type contributors (example)
Welcome aboard! I for one am quite excited about rtika. We'd also love a blog post about your package, either a short-form intro to it (https://ropensci.org/tech-notes/) or long-form post with more narrative about its development. ((https://ropensci.org/blog/). If you are interested in writing one, let us know and, @stefaniebutland will be in touch about content and timing.
Writing a long form post sounds good. It's an opportunity for me to introduce the package and reflect some on the process. Please express my interest and thanks for the opportunity to @stefaniebutland .
Okay, regarding the logistics of the transfer, it's now at:
https://github.com/ropensci/rtika
That was super easy. The README links are also changed, yet I don't know how to change the github repository title blurb at the very top to:
R Interface to Apache Tika https://ropensci.github.io/rtika/
I guess I no longer control repository level settings, which is okay, but that link should be changed.
I made that change. You should have repo settings control now. All set! I'm going to close this issue, but @stefaniebutland will respond here about blog timing. Here are our editorial and technical guidelines for posts: https://github.com/ropensci/roweb2#contributing-a-blog-posts, and some other posts in the onboarding series https://ropensci.org/tags/review/
👍
Hi @predict-r. Great to hear that you are interested in contributing a blog post and thanks to Noam for providing that link to recommendations. Would you be able to submit a draft via pull request by March 27? That will give us an opportunity to review it, give you feedback and set a date for publication.
Greetings @stefaniebutland. That is reasonable. I'll submit the draft by the 27th. Based on https://github.com/ropensci/roweb2#contributing-a-blog-posts, I know what to try. If there are any examples of the blog you particularly liked or want to recommend, let me know and I'll take a closer look. Thanks.
@predict-r This will give you a range of examples https://ropensci.org/tags/review/. Browsing them will give you an idea of the length you prefer, and different angles on the narrative.
I'm happy to give feedback on an early version or outline if that helps you.
@stefaniebutland, I posted a draft here:
https://github.com/predict-r/roweb2/pull/1
It's a pull request on a fork...hope I did that correctly.
Thank you for doing this @predict-r.
hope I did that correctly It looks like you made a pull request to your own fork of our repo.
From https://github.com/predict-r/roweb2 you should create a pull request with branch predict-r-patch-1 in your fork to master on the ropensci repo. It will look like the image here

I have a few other blog posts to review first so I will get you some feedback likely early next week.
Summary
Extract text or metadata from over a thousand file types. This is an R interface to Apache Tika, a content and metadata extraction tool. Tika is written in Java and is maintained by the Apache Software Foundation since 2007.
https://github.com/predict-r/rtika
It fits into data extraction, especially text extraction. This package helps extract either a plain text or XHTML rendition of most common file types, without leaving R. It includes parsers for Microsoft Office (both the old and new formats), OpenOffice, iWorks, WordPerfect, PDF, ePub, rtf, HTML, XML, and Mbox email.
The hours of labor spent on these parsers is unbelievable. I recall Tika was intended to prevent duplicate work at Apache, and has since became a foundational library for Apache search engine projects.
Tika also detects metadata from over a thousand file formats. These include basic features like Content-Encoding and mime type sniffing. Beyond that, there are specialized extractors for scientific formats (ISA-Tab, Matlab), geospatial formats (GDAL, ISO-19139), image Exif data from tiff or jpeg, and many more.
The long list of formats is here: https://tika.apache.org/1.17/formats.html
Computational social scientists, digital humanities scholars, historians, or any person that works with large digital archives filled with possibly unpredictable file formats.
For example, consider a social scientist studying the California state legislature, as I am. The legislature publishes a database that includes, among other things, Microsoft Word documents created over the past decades. These are at http://leginfo.legislature.ca.gov/.
I've used Tika to automatically extract text from decades of Word documents, even if I don't know the particular version of Word beforehand. Further, with the XHTML output option, I can preserve tables inside each Word document.
There are specialized parsers for several of the file types Tika processes, such as antiword for older Microsoft Word documents. Overlap is going to be inevitable. I have used antiword and it worked well, except that some of my files did not have a file type affix (e.g. .docx or .doc), and could be in either format, and so it took some trial and error to get it working. It turned out antiword does not process the modern XML based Word formats. It also does not render the Word document into XHTML, which could preserve tables from the Word document.
Listing all the document parsers that Tika overlaps with would take an enormous amount of space, since it processes over a thousand file types. In general, however, Tika's aims to streamline text extraction within large archives. Archives often contain heterogeneous and unpredictable file formats. Tika handles most of what I throw at it.
The other package I found that interfaces with Apache Tika is tikaR, released in September of 2017 (See: https://github.com/kyusque/tikaR). It provides only XHMTL renditions of documents, and uses the rJava package to interface with Tika.
I was a Java developer and implemented a rJava interface with Tika in 2012, when I was considering my first package for R (the project is empty, but I set up a repository on r-forge back then: https://r-forge.r-project.org/projects/r-tika/). My rJava code worked, but I concluded at the time that the Apache Tika command line interface (CLI) was sufficient for myself and most people. I ended up just crafting some specialized system calls to the command line for my own work. The r-forge repository remains empty because I didn't want to maintain a complex Java codebase.
Recently, the Tika project came out with a very nice batch processor in their command line that I found to be the most efficient method to handle tens of thousands of documents. It was too good to keep to myself.
Unlike the rJava functions used by tikaR, I essentially send Tika a file list to process, and then craft a specialized call to the command line to invoke the batch processor. Retrieving the converted results is pretty straightforward. The code is simple and easy to maintain.
Unlike tikaR, I've included a variety of tests and keep track of test coverage, and anticipate that maintaining this package will be fairly simple, even without Java knowledge. There are also more features implemented in my package than tikaR, such as the ability to use multiple threads.
Requirements
Confirm each of the following by checking the box. This package:
Publication options
paper.mdmatching JOSS's requirements with a high-level description in the package root or ininst/.Detail
[x] Does
R CMD check(ordevtools::check()) succeed? Paste and describe any errors or warnings:[x] Does the package conform to rOpenSci packaging guidelines? Please describe any exceptions:
If this is a resubmission following rejection, please explain the change in circumstances:
If possible, please provide recommendations of reviewers - those with experience with similar packages and/or likely users of your package - and their GitHub user names:
Jeroen Ooms has worked on text file parsers and I respect his work immensely.