thanks for your submission @aaronwolen
@maelle has been assigned as editor
Closed aaronwolen closed 4 years ago
thanks for your submission @aaronwolen
@maelle has been assigned as editor
Thanks for your submission @aaronwolen! Here are a few questions/comments before I can start looking for reviewers.
The installation instructions should include information about the authentication, and make it clear a token will be needed. Moreover, in a CONTRIBUTING.md file you could explain what scope a token needs for testing the package. I created one with scope for everything. :grin:
The repo also lacks a code of conduct see https://ropensci.github.io/dev_guide/collaboration.html#friendlyfiles
Two tests fail
── Test failures ────────────────────────────────────────────────────────── testthat ────
> library(testthat)
> library(osfr)
Automatically registered OSF personal access token
>
> test_check("osfr")
── 1. Error: (unknown) (@test-osf_ls.R#5) ──────────────────────────────────────────────
Not found.
HTTP status code 404.
1: osf_retrieve_node("brfza") at testthat/test-osf_ls.R:5
2: raise_error(out)
3: http_error(x$status_code, x$errors[[1]]$detail)
4: stop(args, msg, call. = FALSE)
── 2. Error: (unknown) (@test-osf_tbl.R#37) ────────────────────────────────────────────
Not found.
HTTP status code 404.
1: osf_retrieve_node("brfza") at testthat/test-osf_tbl.R:37
2: raise_error(out)
3: http_error(x$status_code, x$errors[[1]]$detail)
4: stop(args, msg, call. = FALSE)
══ testthat results ════════════════════════════════════════════════════════════════════
OK: 90 SKIPPED: 1 FAILED: 2
1. Error: (unknown) (@test-osf_ls.R#5)
2. Error: (unknown) (@test-osf_tbl.R#37)
Error: testthat unit tests failed
Execution halted
1 error ✖ | 0 warnings ✔ | 0 notes ✔I have not looked in detail, is it because this is a node that only exists in the account you use for testing? Couldn't the node be created before testing so that anyone can run the tests?
goodpractice::gp() mentions a few too long code lines.
I see there are a few open issues in your repo, labelled "feature requests". Are these features you plan to implement in the near future? In particular, before reviews?
Reviewers: @HeidiSeibold @cboettig Due date: 2018-02-20
Hi, @maelle! Thanks for checking out the package and I apologize for the delayed response—I’ve been out traveling most of this week and am still playing catchup.
I made the following changes based on your comments:
reformatted many of the overly long lines (centerforopenscience/osfr@eb9a81b)
(There are still a few places where I’m breaking the 80 line rule, primarily for errors, warnings, and other messages that aren’t as easily split up.)
Regarding the test failures you encountered, I believe the problem was the tests were run on the main OSF server (osf.io) and several of the required resources are located on OSF’s testing server (test.osf.io), which is where COS prefers development take place. The contributing guidelines mention you need to set OSF_SERVER=test to switch to the testing server, but it’s easy to miss.
This really underscored that the testing infrastructure was a little fragile, so I did some restructuring and added checks to skip certain tests if they’re being run on the production server or if no PAT is defined (centerforopenscience/osfr@81cfe71). It was a big enough change that I bumped the version to 0.2.2.
Thanks again for looking it over and please let me know if you have any other questions or follow-up comments.
Thanks @aaronwolen, great work! Sorry for missing the contributing guidelines! Two small suggestions:
"Create a .Renviron file in the root of your project directory" or one could edit the existing general .Renviron/create one (that's what I did :-) ). You could mention the handy usethis::edit_r_environ() function for that.
Regarding the long lines, you could add # nolint at the end of the ones you do not want to split cf https://github.com/jimhester/lintr#project-configuration
I'll now look for reviewers! :smile_cat:
Great suggestions! I did not know about edit_r_environ()—that package is just chock full of goodness—I’ll be sure to mention it.
@aaronwolen Great definition of usethis :grin:
@HeidiSeibold @cboettig Thanks for accepting to review this package! :smile_cat: Your reviews are due on 2019-02-20. As a reminder here's our guide for reviewers and the review template.
@maelle, a new version of purrr was just released that, unfortunately, introduced a bug in osfr. It's fixed in the dev branch but I wanted to verify it's okay to merge the new version now that reviewers have been assigned?
Congrats on fixing that bug so quickly! Yes I think it's fine, I don't think they started reviewing yet (feel free to disagree, @heidiseibold and @cboettig). In general we prefer a stable version but bug fixes are a good reason to change stuff. :smile_cat:
I will just add my review template below, so you know what I am up to :smile_cat:
Please check off boxes as applicable, and elaborate in comments below. Your review is not limited to these topics, as described in the reviewer guide
The package includes all the following forms of documentation:
URL, BugReports and Maintainer (which may be autogenerated via Authors@R).Estimated hours spent reviewing: 10
I have limited expertise on how to correctly test functionality of a package which talks to a server, so I checked the Automated tests-Box, but just because I could not see any obvious problems and the check ran through. If you need an expert here, I could ask if @giuseppec can take a look.
Documentation on osf_upload() says:
overwrite Logical, overwrite an existing file with the same name (default FALSE)? If TRUE, OSF will automatically update the file and record the previous version. What happens for FALSE? Does it not upload at all or does it upload as a separate file?
The error message does not really help me solve the problem. Why does the authentication say it is ok, but then it does not work?
> osf_auth(token = "<mytoken>")
Registered PAT from the provided token
> osf_retrieve_node("vk95n")
Error: User provided an invalid OAuth2 access token
HTTP status code 401. (Btw. the issue for me was that I was using the token from the test server instead of the actual osf, where the project lives)
How do I list all components within a project?
For one of my projects I do not receive all components, when I ask for it (e.g. talk_2015_07_user, compare code to screenshot).
> proj <- osf_retrieve_node("vk95n")
> osf_ls_nodes(proj)
# A tibble: 10 x 3
name id meta
<chr> <chr> <list>
1 talk_2017_09_novartis rknwh <list [3]>
2 talk_2017_10_advisory_board 7uj8p <list [3]>
3 talk_2017_11_biogen 8tzr6 <list [3]>
4 talk_2017_11_helmholtz f5bnu <list [3]>
5 talk_2018_01_institutskolloquium n26z5 <list [3]>
6 talk_2018_03_defense gqzrf <list [3]>
7 talk_2018_07_ecda azcqx <list [3]>
8 talk_2019_01_KI_med 3j4a8 <list [3]>
9 talk_2015_07_isnps e93gh <list [3]>
10 talk_2015_06_iroes 9npxe <list [3]>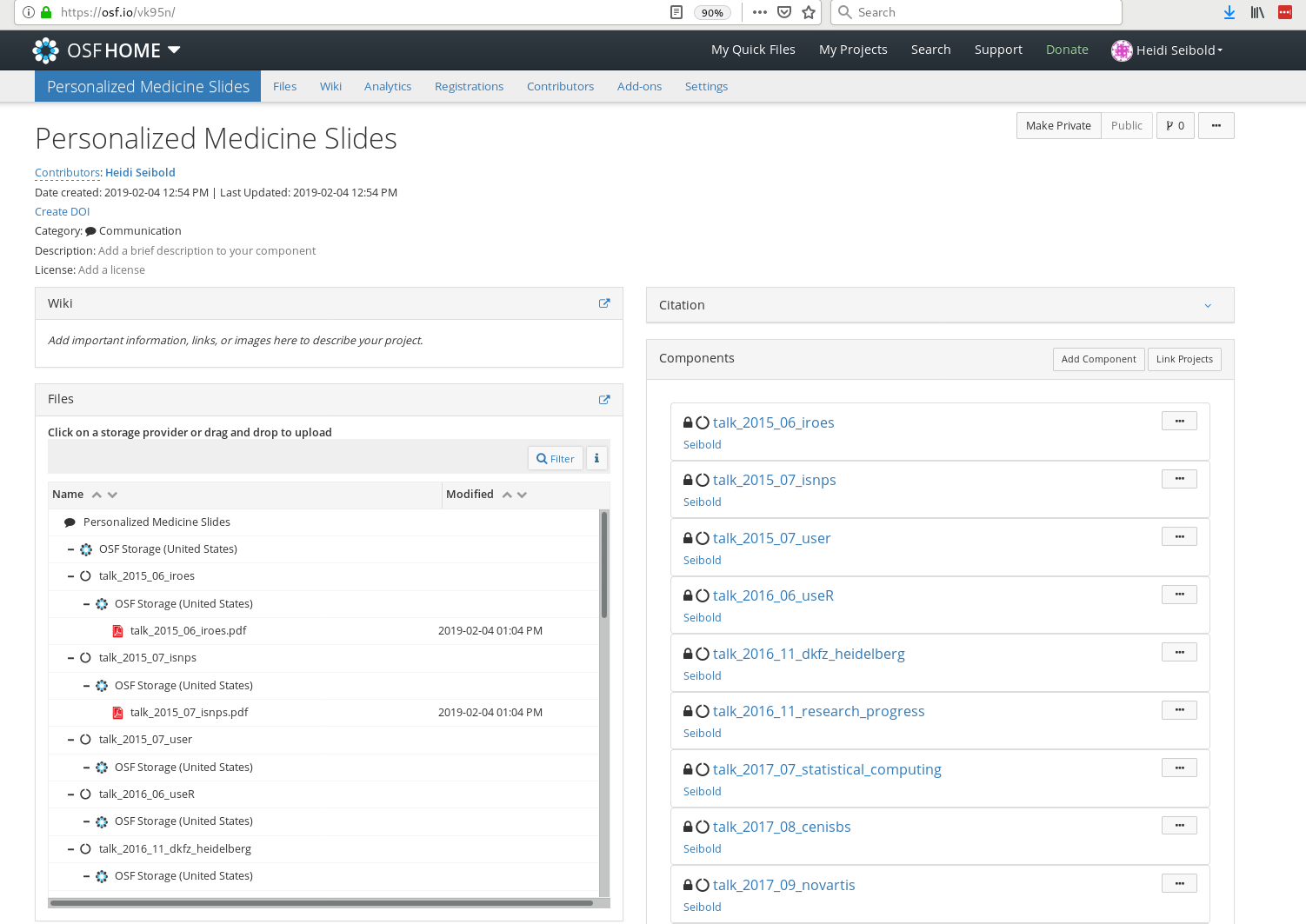
https://osf.io/vk95n/ Any idea what is causing this?
If I understand correctly, your main object in the package is osf_tbl_node, which consists of name, id and meta. I would appreciate some more documentation on the meta part. What info does it contain?
meta would also contain the info on the components of the project, but this seems to not be the case. osf_tblid might be helpful for many users.What is the difference between a project, a component and a directory? I think I have a rough understanding of it, but it might be helpful to write this down somewhere explicitly.
Will osfr in the future also allow interacting with files on e.g. GitHub, which are connected to osf?
(Related to above question, but more of a personal interest question: if I have files connected to osf from GitHub, would the files be lost if GitHub was shut down?)
Are you planing on creating a pkgdown-website?
@aaronwolen if you have time, I would appreciate you already answering my current questions. This way it will be easier for me to move forward. Thanks :)
@HeidiSeibold live-updated review, that's open science for sure. :wink:
Question: what is the difference between:
remotes::install_github()
and
devtools::install_github()
The package README is instructing to use the former. Does that matter at all @maelle?
@richfitz said "devtools::install_github calls remotes::install_github in recent devtools versions (since 2.0.0)" so there shouldn't be any difference. :-)
I am trying to build the package on my computer (Debian):
hseibold(08:32)~/Documents/git$ git clone git@github.com:CenterForOpenScience/osfr.git
Cloning into 'osfr'...
remote: Enumerating objects: 127, done.
remote: Counting objects: 100% (127/127), done.
remote: Compressing objects: 100% (79/79), done.
remote: Total 4223 (delta 76), reused 76 (delta 45), pack-reused 4096
Receiving objects: 100% (4223/4223), 1.15 MiB | 2.46 MiB/s, done.
Resolving deltas: 100% (2810/2810), done.
hseibold(08:33)~/Documents/git$ cd osfr/
hseibold(08:33)~/Documents/git/osfr (master)$ R CMD build .
* checking for file ‘./DESCRIPTION’ ... OK
* preparing ‘osfr’:
* checking DESCRIPTION meta-information ... OK
* installing the package to build vignettes
* creating vignettes ... ERROR
Warning in engine$weave(file, quiet = quiet, encoding = enc) :
Pandoc (>= 1.12.3) and/or pandoc-citeproc not available. Falling back to R Markdown v1.
Warning in engine$weave(file, quiet = quiet, encoding = enc) :
Pandoc (>= 1.12.3) and/or pandoc-citeproc not available. Falling back to R Markdown v1.
Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
Quitting from lines 48-50 (getting_started.Rmd)
Error: processing vignette 'getting_started.Rmd' failed with diagnostics:
Authentication credentials were not provided.
HTTP status code 401.
Execution haltedThe pandoc issue is probably something I have to fix.
How do you solve the authentication issue @aaronwolen?
The reason why I wanted to build it myself was to have a nicely viewable Vignette, which I apparently don't get if I install from GitHub.
> browseVignettes(package = "osfr")
No vignettes found by browseVignettes(package = "osfr")@HeidiSeibold I had the same authentication issues before @aaronwolen pointed me to https://github.com/CenterForOpenScience/osfr/blob/master/.github/CONTRIBUTING.md :-)
Hi, @HeidiSeibold! I just noticed your edits and responded below:
- Documentation on
osf_upload()says:overwrite Logical, overwrite an existing file with the same name (default FALSE)? If TRUE, OSF will automatically update the file and record the previous version.What happens for
FALSE? Does it not upload at all or does it upload as a separate file?
The default behavior is to throw the following error
Error: File already exists at destination
* Set `overwrite=TRUE` to upload a new version(Btw. the issue for me was that I was using the token from the test server instead of the actual osf, where the project lives)
I've run into this several times myself. I'm considering adding a startup message that informs the user when OSF_SERVER is defined and they're running on the test server.
- How do I list all components within a project?
The n_max argument determines how many components will be listed (really just limiting the number of calls made to the API). The default is 10 but setting it to Inf will return all components.
I see now that this behavior is not documented! I'll improve the documentation for this argument in the dev branch. Thanks for pointing that out!
Thanks for the questions! I'll keep checking back for more.
@maelle I have trouble checking
and
because for many examples it does not make sense to run them without modification. I will just check them anyway because I can use and modify the examples so it works for me.
@HeidiSeibold I'm not sure I understand your problem?
Sorry, I'll try to clarify:
Many examples should not be run, because they would upload data to osf and you don't want the same data to be uploaded to osf every time someone tries the package. So there is a dont-run-example, which I modify and then try, so I actually upload useful things to osf.
Same for the vignette.
Thanks for the additional comments, @HeidiSeibold!
- For one of my projects I do not receive all components, when I ask for it (e.g.
talk_2015_07_user, compare code to screenshot).
I wanted to make sure you saw my previous comment that mentions using the n_max argument to specify the max number of results returned by osf_ls_nodes(). I updated the docs to indicate that n_max = Inf will retrieve all results for a query (CenterForOpenScience/osfr@8ff84f4b).
I believe this is related to your specific example regarding OSF project https://osf.io/vk95n/. The default value for n_max is 10, so that's likely why the component talk_2015_07_user was not included in the results. Try setting n_max to something > 10 or Inf and let me know if that resolves your issue. BTW, the screenshot shows only 9 components and the project components are private, so I'm unable to verify myself.
If I understand correctly, your main object in the package is
osf_tbl_node, which consists ofname,idandmeta. I would appreciate some more documentation on themetapart. What info does it contain?
- I assumed that
metawould also contain the info on the components of the project, but this seems to not be the case.- I would like to see a deeper documentation of
osf_tbl- Maybe also a note on the connection between the weblink and the
idmight be helpful for many users.
We do provide some documentation in ?osf_tbl, and mention that meta contains "a list-column that stores the processed response returned by OSF's API".
I totally agree it could be more detailed. Please let me know if you have any specific details in mind that you feel are missing, it would really help to highlight some of the areas I'm likely overlooking. For example, your request to note the connection between OSF URLs and ids was great—definitely an oversight on our part. I tried to make it clearer here: (CenterForOpenScience/osfr@22e8af2)
- What is the difference between a project, a component and a directory? I think I have a rough understanding of it, but it might be helpful to write this down somewhere explicitly.
Projects, components, and directories represent the 3 different levels of organization on OSF that can be used to structure a project. OSF actually provides some really great documentation on how these entities compare to each other:
We reference these links in a few places but they should probably be more front and center in the README, or perhaps in, ?osfr. That might help users less familiar with OSF get started.
- Will
osfrin the future also allow interacting with files on e.g. GitHub, which are connected to osf?
Yes! We're definitely planning to add support for the other providers that OSF works with. The plan was to first make sure the package works well/reliably with OSF's native cloud storage, and then include 3rd-part services in a future release.
(Related to above question, but more of a personal interest question: if I have files connected to osf from GitHub, would the files be lost if GitHub was shut down?)
Adding GitHub to an OSF project just creates a connection between the two services, but the GitHub files remain on GitHub. So if GitHub shuts down, the files would be lost.
You can backup external files stored on a connected 3rd party service by registering the project.
- Are you planing on creating a pkgdown-website?
Planned and deployed: https://centerforopenscience.github.io/osfr :slightly_smiling_face:
Thanks @aaronwolen for your answers. They are very helpful and I hope I am helpful in making your package even better than it is already :wink:
For one of my projects I do not receive all components, when I ask for it (e.g. talk_2015_07_user , compare code to screenshot).
I wanted to make sure you saw my previous comment that mentions using the n_max argument to specify the max number of results returned by osf_ls_nodes(). I updated the docs to indicate that n_max = Inf will retrieve all results for a query (CenterForOpenScience/osfr@8ff84f4).
Sorry, yes I did read it but immediately forgot. Works now!
We do provide some documentation in ?osf_tbl, and mention that meta contains "a list-column that stores the processed response returned by OSF's API".
I totally agree it could be more detailed. Please let me know if you have any specific details in mind that you feel are missing, it would really help to highlight some of the areas I'm likely overlooking. For example, your request to note the connection between OSF URLs and ids was great—definitely an oversight on our part. I tried to make it clearer here: (CenterForOpenScience/osfr@22e8af2)
Yeah I do agree that a bit more detail would be helpful. Some examples:
meta always have only one list item data?attributeslinksrelationshipsmeta/dataWe reference these links in a few places but they should probably be more front and center in the README, or perhaps in, ?osfr. That might help users less familiar with OSF get started.
Yes I agree. These objects are essential for understanding how to interact with OSF. Also an overview on actions which can be done with the objects would be helpful. On OpenML we are using a cheatsheet with clear visualisations to help users find the functionality they need at each moment. I think OSF has similar features (listing, downloading, etc) so this might be helpful to look at.
Thanks again, @HeidiSeibold. I'll be sure to include more details about the meta column in the next release. We're also planning to add a function that allows users to easily retrieve useful information about OSF objects from meta but I'm still working on the implementation.
I also think a cheatsheet is a great idea, it's definitely on the todo list. The OpenML cheatsheet is a really nice example, thanks for sharing!
:wave: @cboettig, friendly reminder that your review is due today. 😸
sorry I'm late! 😊
Please check off boxes as applicable, and elaborate in comments below. Your review is not limited to these topics, as described in the reviewer guide
The package includes all the following forms of documentation:
URL, BugReports and Maintainer (which may be autogenerated via Authors@R).Estimated hours spent reviewing: 4.5
Really professionally developed package here. All the tests and examples ran smoothly for me. The documentation, the unit tests, and the code were easy to ready and understand and look very thoughtfully implemented. I have a few minor comments, some conceptual questions about the design, and some recommendations on regarding the documentation.
I think this is a very valuable package and am looking forward to testing it out in real use cases in my lab group over the coming weeks, since I think it could serve many of our needs that aren't met by other existing utilities. exciting stuff!
Are there any operations from the API that can be supported without authentication? In particular, I think the most useful feature would be the ability to download public files without an API key. These can be downloaded in the browser without logging in, so this would seem comparable. (Unfortunately from the browser end, I don's seem to be able to get a hard URL for the download since the browser interface itself doesn't seem to be RESTful...)
While I value the low-level API, manually recreating directory structure in the create_project example becomes tedious for a large(r) project. Rather than indicate the names of each file and folder, it would be very nice to have a higher level interface that could simply import the files and folders of an existing project; potentially indicating which files to exclude (a la .gitignore). This would be straight forward enough for a user to implement with the existing package, but would make more sense at this level.
Similarly, I imagine some users might want to import a project from GitHub. I saw http://help.osf.io/m/addons/l/837075-connect-github-to-a-project, but would be nice to see a scripted way to do this from osfr if possible; at least after a one-time online enable.
While this may be beyond what the API can provide, I think it would also be really nice to be able to control user permissions from the command line. In particular, it would be nice to be able to grant other users permission to view or view & edit files automatically from the R package by specifying user IDs. (For instance, this can be done in DataONE, though the downside to their system imho is the shortlived tokens make daily use for private / in development work inpractical.)
Are there plans to extend package functionality beyond the current scope of just file/folder/node management? The web interface even without extensions is so much richer that this currently feels somewhat narrow, (though is certainly the most useful bit.) For instance, it might be nice to get download statistics etc for a project or file.
.token in gh package, also see usethis & devtools examples), giving the user an alternative to setting the environmental variable. This can be useful for more complex authentication designs (scripts authenticating in multiple users) or for developers, or just more convienient for a one-off function call. README.Rmd/md:
Why Sys.unsetenv(c("OSF_SERVER", "OSF_PAT"))?
r_project <- osf_retrieve_node("e81xl") 404 error: invalid token (with & without my token). Presumably not meant to be run with test account tokens?
Consider linking to the (very nice) vignettes directly.
very nice CONTRIBUTING.md, minor tweaks to wording in a PR!
Thanks @cboettig!😸
@heidiseibold are you done with your review? If so please update the "Estimated hours spent reviewing:" field. 🙂
Thanks for the comments @cboettig.
Are there any operations from the API that can be supported without authentication? In particular, I think the most useful feature would be the ability to download public files without an API key. These can be downloaded in the browser without logging in, so this would seem comparable. (Unfortunately from the browser end, I don's seem to be able to get a hard URL for the download since the browser interface itself doesn't seem to be RESTful...)
You should only need authentication to make edits to projects, or to download private files. Any public node or file can be accessed without authentication. So osf_retrieve_file('fgjvw') %>% osf_download should work without a key. Are you getting an error, or do we just need to make it more clear in the documentation what can be done without authentication?
Similarly, I imagine some users might want to import a project from GitHub. I saw http://help.osf.io/m/addons/l/837075-connect-github-to-a-project, but would be nice to see a scripted way to do this from
osfrif possible; at least after a one-time online enable.
You is technically possible through the API, but those endpoints aren't particularly stable right now. We also decided to restrict the initial release of the package to just work just with OSF storage, and potentially tackle add-ons in future releases if there is community requests for this.
- While this may be beyond what the API can provide, I think it would also be really nice to be able to control user permissions from the command line. In particular, it would be nice to be able to grant other users permission to view or view & edit files automatically from the R package by specifying user IDs. (For instance, this can be done in DataONE, though the downside to their system imho is the shortlived tokens make daily use for private / in development work inpractical.)
- Are there plans to extend package functionality beyond the current scope of just file/folder/node management? The web interface even without extensions is so much richer that this currently feels somewhat narrow, (though is certainly the most useful bit.) For instance, it might be nice to get download statistics etc for a project or file.
These two point are related. It's possible to do the first point through the API. The API is really extensive, and as you correctly pointed out enables much richer functionality than the package currently covers. I think adding contributor management is a great suggestion for a future version of the package, and we do have plans to extend out the functionality in later releases. Given the long development history fo this package, and our desire to have a stable version of the package released and on CRAN, we decided to focus in on a smaller set of core features to start in order to limit the initial scope to something manageable for the first official version of the the package. For future versions, I know we have a few features in the back of our minds that we'd like to include, and are also very open to community feedback about what features would be most useful to them to help prioritize when goes out in future releases.
r_project <- osf_retrieve_node("e81xl")404 error: invalid token (with & without my token). Presumably not meant to be run with test account tokens?
I'm not able to recreate this error. Were you by chance pointing towards the test.osf.io API endpoint, rather than the main osf.io API when you made that call? The 'usage example' and the 'vignette' use example public projects form OSF, rather than our test server.
Re 404 error: thanks, yes, I'm able to get this to work in a fresh session where I have not set OSF_SERVER=test in .Renviron. I am not able to get it to work though in the session where I have set test in .Renviron, even after I run Sys.unsetenv(c("OSF_SERVER", "OSF_PAT")) though, which I would have thought would be sufficient. Can you see if you get a 404 if you set test in the .Renviron but then unload it?
Aside from this weird technical glitch though, this is great, very nice to be able to download public files without a token!
I still think it would be nice to support creation of a project that has multiple files and folders as a one-liner function call, rather than the very manual current example. This task is the feature-example shown in the readme and getting started docs, but takes 5 function calls to upload one data file in one project. I'd love something more analogous to git init, git add -A/git commit, git push style to upload a larger project with many files. This could be built out entirely on the R end with just a bit of dir.list() or fs::dir_ls() style commands and would be helpful in getting larger projects started. Automating the process for many files is probably one of the more compelling reasons to create a package manually in the first place instead of using the web interface, and I'd love to review such a capability here. Nevertheless, I appreciate the goal of not holding up your submission to CRAN so I'd be happy to see these things just on the roadmap / milestones; clearly the core functionality is both solid and valuable as is, so this is just a suggestion which I think would improve the first-impressions of usability for real-life cases.
First of all, thanks so much for the thoughtful review and kind words, @cboettig. It’s great to hear that osfr might be useful in your lab.
Courtney did a nice job addressing your questions concerning authentication and supporting add-ons, so I’ll start with your suggestion about adding a higher level interface for adding an existing project to OSF:
While I value the low-level API, manually recreating directory structure in the
create_projectexample becomes tedious for a large(r) project. Rather than indicate the names of each file and folder, it would be very nice to have a higher level interface that could simply import the files and folders of an existing project; potentially indicating which files to exclude (a la.gitignore). This would be straight forward enough for a user to implement with the existing package, but would make more sense at this level.
I totally agree the number of steps required for certain tasks is tedious at the moment. Now that we have a core set of atomic functions for performing (what I assume are) the most common OSF operations, I plan to focus on making typical workflows more convenient. Your example of uploading/importing an existing project to OSF is a perfect example.
I’ve been working on updated versions of the upload and download functions that support multiple files/directories. This should make the process of adding an existing project to OSF slightly less painful since you could upload your entire project directory at once:
my_proj <- osf_create_project("My Existing Project")
osf_upload(my_proj, path = getwd())Love your suggestion for a .gitignore-style feature! It would be a great complement to the above workflow, making it possible to implement specific filters without requiring any additional arguments that might complicate the interface.
Do you think the combination of these features would do the trick?
Are there plans to extend package functionality beyond the current scope of just file/folder/node management? The web interface even without extensions is so much richer that this currently feels somewhat narrow, (though is certainly the most useful bit.) For instance, it might be nice to get download statistics etc for a project or file.
In terms of providing access to the metadata that OSF provides about projects and files, that’s absolutely on the roadmap. Much of that info is already present within the meta column, so a short-term fix will be to better document what’s hiding in there (as suggested by @HeidiSeibold), more advanced users can pluck out what they need. After that, I’m planning to include a new function for adding relevant attributes to an osf_tbl. Here’s an example:
psych_rp <- osf_retrieve_node("ezum7")
psych_rp %>%
osf_get(c("category", "date_created", "tags"))
# # A tibble: 1 x 6
# name id category date_created tags meta
# <chr> <chr> <chr> <dttm> <list> <list>
# 1 Estimating the Reproducibility of Psychological Science ezum7 communication 2015-07-20 17:42:16 <list [3]> <list [3]>This will also coincide with a new ls-like argument for osf_ls_files() and osf_ls_nodes() to include more information in their output, e.g.
osf_ls_files(psych_rp, info = "extra")
# # A tibble: 10 x 7
# name id kind size modified downloads meta
# <chr> <chr> <chr> <chr> <dttm> <int> <list>
# 1 RPP_Supplement.pdf 55f1a8d18c5e4a093bc70500 file 822.89 kB 2015-09-18 01:11:16 517 <list [3]>
# 2 RPP_SI_Figures 55ad3c7e8c5e4a527e115b8d folder 0 B 2015-09-18 01:11:16 0 <list [3]>
# 3 RPP_Appendices.pdf 55ad337b8c5e4a5277114f47 file 188.72 kB 2015-09-18 01:11:16 285 <list [3]>
# 4 RPP_Figure3.pdf 55ad3c0b8c5e4a527c115553 file 27.83 kB 2015-09-18 01:11:16 130 <list [3]>
# 5 RPP_Figure1.pdf 55ad3c0b8c5e4a527e115b7b file 16.59 kB 2015-09-18 01:11:16 191 <list [3]>
# 6 RPP_Figure2.png 55ad3c0c8c5e4a527e115b7e file 51.27 kB 2015-09-18 01:11:16 124 <list [3]>
# 7 RPP_Table_1.pdf 55ad3c168c5e4a527e115b81 file 31.23 kB 2015-09-18 01:11:16 168 <list [3]>
# 8 RPP_Table_2.pdf 55ad3c178c5e4a527711503c file 43.79 kB 2015-09-18 01:11:16 167 <list [3]>
# 9 RPP_Figure3_compressed.jpg 55e05f118c5e4a2f9b1a72d7 file 75.96 kB 2015-09-18 01:11:16 93 <list [3]>
# 10 RPP_Figure1_panelA_compressed.jpg 55e0b3408c5e4a6d5cadaede file 19.86 kB 2015-09-18 01:11:16 87 <list [3]>The one issue I see is the lack of support to pass a token to the function call. Typical design patterns I like here provide the option of specifying a token as an optional argument (e.g.
.tokeninghpackage, also seeusethis&devtoolsexamples), giving the user an alternative to setting the environmental variable. This can be useful for more complex authentication designs (scripts authenticating in multiple users) or for developers, or just more convenient for a one-off function call.
It was something I considered initially but ultimately decided against, figuring most users would opt to authenticate using the env variable, and those who don't could still pass their PAT directly to osf_auth(). Do you think that would suffice in the multi-user scenarios you're envisioning?
- Why
Sys.unsetenv(c("OSF_SERVER", "OSF_PAT"))?
Code chunks in the README need to access public resources on OSF. Since the README is usually only rendered in a dev environment where OSF_SERVER=test and the OSF_PAT was generated on
As to the 404 you're experiencing, I'm not able to recreate it on macOS running R 3.5.1. Could you give me a little more info your setup?
BTW, happy to keep this dialogue going post-review if you have additional ideas about how to make the package more useful to your lab.
@aaronwolen This is great.
Regarding uploading a project's files & directory structure in one go with
my_proj <- osf_create_project("My Existing Project")
osf_upload(my_proj, path = getwd())that sounds like a most welcome addition! cc'ing @karinorman who has been battle-testing this with some complex project structures, we'll try and give that a test-drive. The previously manual approach was a bit of a show-stopper for us at first pass.
Re 404, I see that also on MacOS (high Sierra) on R 3.5.2; but it seems to be a weird side-effect of using Sys.unsetenv(c("OSF_SERVER", "OSF_PAT")) interactively? Anyway, seems related to the test setup and unlikely to be an issue for users, so probably not worth diving into.
Regarding the token, I'm not quite clear what the workflow would look like if I wanted to say, upload to three different projects with distinct tokens in the same script; I guess just call osf_auth() each time before each function call? Relying on side-effects instead of being able to pass all parameters if need be undercuts a functional-programming workflow, though in practice it's probably not a big deal. I certainly agree that most users will just want to set .Renviron and that the functions should check for the tokens there if no .token argument is given, I just like functions to also be able to run as pure functions where all arguments are specified as arguments. Happy to let that go here.
@maelle Aaron, Courtney and co have satisfied all my issues here, so I'm happy to approve this package :+1: and continue any further discussion with them about full-directory uploads etc in the issues tracker for the osfr project itself instead. Very nice package here and a great contribution to the R community.
Just a quick correction to my comments above: looks like I jumped the gun here and the "upload local directory+files) feature is still under development, correct?
It would be nice to give it a test run and see it show up in the documentation (perhaps in place of the more tedious current example). Happy to test-drive it from a dev branch if you point us in the right direction
That's right. My plan was to cut a minor release (CenterForOpenScience/osfr#97) that includes a few bug fixes and addresses most of the issues raised here so current users can benefit from the improvements asap. After that I'll focus on some of the bigger changes for a v0.3.0 release, since they'll likely introduce some breaking changes.
There is a feature branch that includes experimental support multi-file downloads here. It still needs some polish (e.g., progress bars) but works pretty well at this point. It'd be great to have you (or anyone else reading this) test it out!
Uploading multiples files+directories, however, is still very much a work in progress (i.e., not functional). It's something I would personally use quite a bit in my own work. So it's definitely next on the list, if only for selfish reasons 😄.
Thanks @aaronwolen this sounds great. I think we'd get a lot of use out of multi file uploading and downloading too -- looking forward to it.
Ping @HeidiSeibold cf https://github.com/ropensci/software-review/issues/279#issuecomment-466484075 :smile_cat:
Sorry, yes I am done. :see_no_evil:
:wave: @aaronwolen can you please update this thread once you're done taking reviews into account? Thank you!
Thanks for following-up, @maelle! Apologies for the delay. Since initiating this review I've changed employers and moved to a new state... so it's been a hectic month 😃.
I just pushed v0.2.4 which should include all of the suggested changes with the exception of multi-file uploading and downloading. That's definitely next on the list but will require a little more time to get it polished and ready for release.
Quite an eventful montu indeed, I hope all went well!
So should we put the review on hold until you implement that feature?
Thanks! I'll defer to you as to whether the review should be placed on hold. I suppose it depends on whether successfully completing the review is contingent on adding the feature or not. It's in the pipeline either way.
Yes I think it's better to have the feature in before accepting the package, thank you!
:wave: @aaronwolen! Any update on the new feature? Thanks! :smile_cat:
Hey @maelle, Things are coming along, albeit a little more slowly than expected (my side-project time has been limited recently). So far I've got multi-file downloads working well 👇

Uploading is still a WIP but I hope to have it completed in the near future 🤞.
Cool work and thanks for the update! :smile_cat:
:wave: @aaronwolen, any update? Thanks 🙂
Hi @maelle, getting very close indeed.
Multi-file uploading and downloading now works fine ✅.
However, I'm still tweaking some of the logic for handling file conflicts. In my original submission osf_upload()/osf_download() had only a binary overwrite argument, which just doesn't scale to multiple files, especially if you want to do something like upload (and later update) an entire project directory.
My solution was to drop overwrite in favor of a new conflicts argument that allows you to optionally "overwrite" or "skip" conflicting files (either uploading or downloading), without stopping the entire transfer.
So this has involved quite a bit of refactoring and additional testing.
The changes are all still in a feature branch but I'm planning to merge to the dev branch soon and put out a call on the Slack channel for some brave beta testers to kick the tires.
Thanks @aaronwolen ! This sounds great. standing by for the ping when you're ready for those beta testers.
👋 @aaronwolen! Any progress with the beta testing? 🙂
You know what? You're never going to believe this... but I think it's ready for testing. 😮
So @cboettig and @HeidiSeibold, if you're still around after all this time, I'd love to get some feedback on the new upload/download implementations. In particular, it'd be great to know if the docs are clear and whether the new behavior of osf_upload()/osf_download() matches your expectations.
If you're willing to give it a go, v0.2.5 (which I'm calling the beta) is available on the dev branch.
remotes::install_github("centerforopenscience/osfr", ref = "dev")The most important changes are detailed in the release notes, so that's a good place to start.
I should note there are still some rough edges that I'll address during this beta phase. For example, the messaging in the upload and download functions is a little inconsistent.
Thanks @aaronwolen!
I am busy next 2 weeks, but might be able to fit a little testing the week after. Is that too late?
Not at all, @HeidiSeibold. I'd appreciate your feedback whenever you get a chance.
Since these changes enable osfr to handle (potentially) lots of files at once, I really want to test them out as thoroughly as possible before opening them up to a wider audience.
Any news @aaronwolen @heidiseibold? 🙂
Submitting Author: Aaron Wolen (@aaronwolen)
Repository: https://github.com/CenterForOpenScience/osfr Version submitted: 0.2.1 Editor: TBD
Reviewer 1: TBD
Reviewer 2: TBD
Archive: TBD
Version accepted: TBD
Scope
Please indicate which category or categories from our package fit policies this package falls under: (Please check an appropriate box below. If you are unsure, we suggest you make a pre-submission inquiry.):
Explain how the and why the package falls under these categories (briefly, 1-2 sentences):
osfr provides a suite of functions for interacting with OSF (https://osf.io), a project management repository that supports researchers across their entire project lifecycle. Using osfr and OSF, researchers can create and manage projects, upload and share materials, or explore publicly accessible projects and download the associated files.
Who is the target audience and what are scientific applications of this package?
Any R users who rely on OSF for managing their research or working with collaborators.
Are there other R packages that accomplish the same thing? If so, how does yours differ or meet our criteria for best-in-category?
No.
If you made a pre-submission enquiry, please paste the link to the corresponding issue, forum post, or other discussion, or @tag the editor you contacted.
Technical checks
Confirm each of the following by checking the box. This package:
Publication options
JOSS Options
- [ ] The package has an **obvious research application** according to [JOSS's definition](https://joss.readthedocs.io/en/latest/submitting.html#submission-requirements). - [ ] The package contains a `paper.md` matching [JOSS's requirements](https://joss.readthedocs.io/en/latest/submitting.html#what-should-my-paper-contain) with a high-level description in the package root or in `inst/`. - [ ] The package is deposited in a long-term repository with the DOI: - (*Do not submit your package separately to JOSS*)MEE Options
- [ ] The package is novel and will be of interest to the broad readership of the journal. - [ ] The manuscript describing the package is no longer than 3000 words. - [ ] You intend to archive the code for the package in a long-term repository which meets the requirements of the journal (see [MEE's Policy on Publishing Code](http://besjournals.onlinelibrary.wiley.com/hub/journal/10.1111/(ISSN)2041-210X/journal-resources/policy-on-publishing-code.html)) - (*Scope: Do consider MEE's [Aims and Scope](http://besjournals.onlinelibrary.wiley.com/hub/journal/10.1111/(ISSN)2041-210X/aims-and-scope/read-full-aims-and-scope.html) for your manuscript. We make no guarantee that your manuscript will be within MEE scope.*) - (*Although not required, we strongly recommend having a full manuscript prepared when you submit here.*) - (*Please do not submit your package separately to Methods in Ecology and Evolution*)Code of conduct