I would be most interested in this as well.
On Sun, Mar 1, 2015 at 11:10 AM, Roger D. Peng notifications@github.com wrote:
One thing I've always wanted is a 'diff' type output for datasets (let's say tabular datasets for now). When I use git to manage projects, changes to the datasets I use are difficult to visualize using the standard diff output, which is line based. That works when rows are changed but not when columns are added/deleted or transformations are made. Is there a way to categorize the types of changes that can be made to a dataset and then visualize them in a useful way?
— Reply to this email directly or view it on GitHub https://github.com/ropensci/unconf/issues/19.
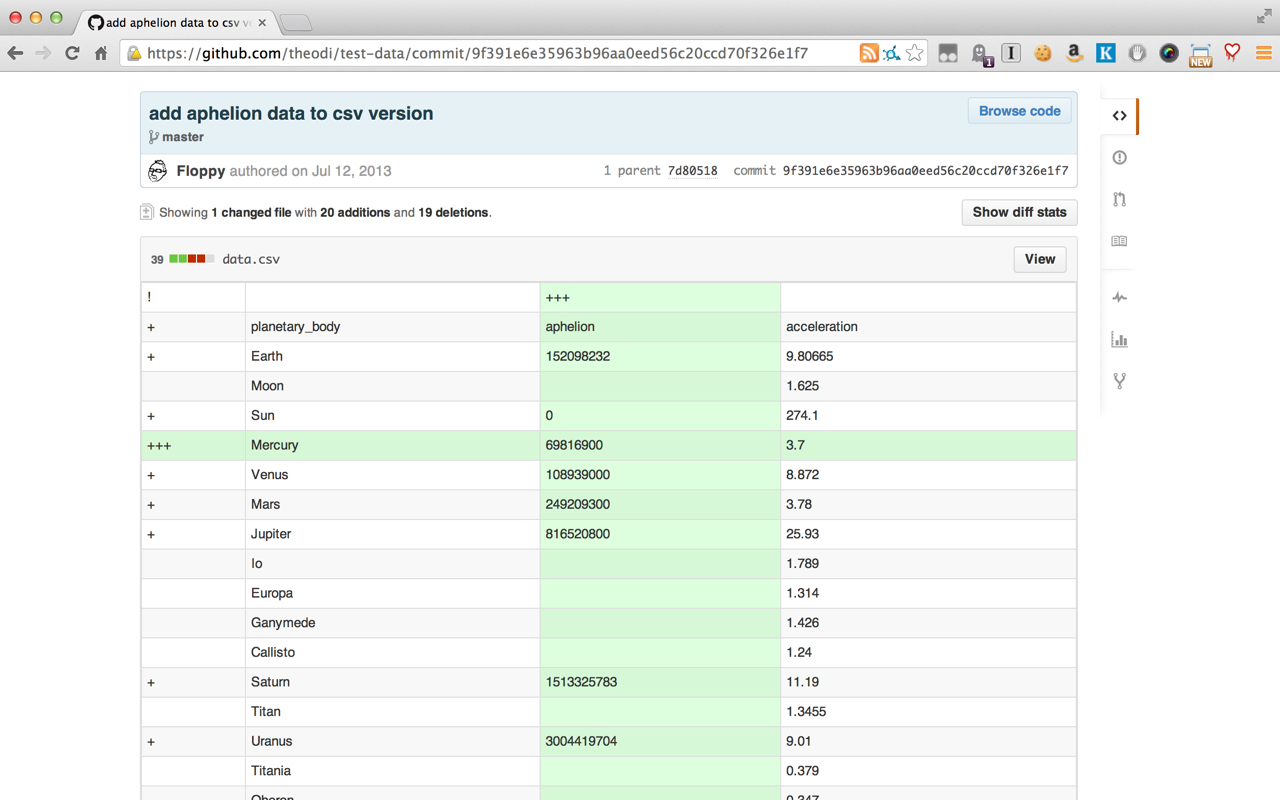










One thing I've always wanted is a 'diff' type output for datasets (let's say tabular datasets for now). When I use git to manage projects, changes to the datasets I use are difficult to visualize using the standard diff output, which is line based. That works when rows are changed but not when columns are added/deleted or transformations are made. Is there a way to categorize the types of changes that can be made to a dataset and then visualize them in a useful way?